High throughput automated analysis of big flow cytometry data
- PMID: 29287915
- PMCID: PMC5815930
- DOI: 10.1016/j.ymeth.2017.12.015
High throughput automated analysis of big flow cytometry data
Abstract
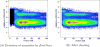
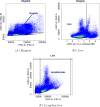
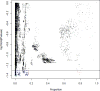
The rapid expansion of flow cytometry applications has outpaced the functionality of traditional manual analysis tools used to interpret flow cytometry data. Scientists are faced with the daunting prospect of manually identifying interesting cell populations in 50-dimensional datasets, equalling the complexity previously only reached in mass cytometry. Data can no longer be analyzed or interpreted fully by manual approaches. While automated gating has been the focus of intense efforts, there are many significant additional steps to the analytical pipeline (e.g., cleaning the raw files, event outlier detection, extracting immunophenotypes). We review the components of a customized automated analysis pipeline that can be generally applied to large scale flow cytometry data. We demonstrate these methodologies on data collected by the International Mouse Phenotyping Consortium (IMPC).
Keywords: Automated analysis; Bioinformatics; Flow cytometry.
Crown Copyright © 2017. Published by Elsevier Inc. All rights reserved.
Figures


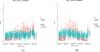











References
-
- Coppola M, Dolecki MN. [2017-09-27];BD Launches BD FACSymphony™ High-Speed Cell Analyzer to Enable Identification and Analysis of Rare Cell Types. 2016 http://www.bd.com/en-us/company/news-and-media/press-releases/feb-8-2016....
-
- Roederer M, Brenchley J, Betts M, De Rosa S. Flow cytometric analysis of vaccine responses: how many colors are enough? Clinical Immunology. 2004;110(3):199–205. - PubMed
-
- [2017-09-11];The Adult and Embryonic Phenotype Pipeline. http://www.mousephenotype.org/impress.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

