Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12Rβ1
- PMID: 29287995
- PMCID: PMC5773378
- DOI: 10.1016/j.immuni.2017.12.008
Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12Rβ1
Abstract
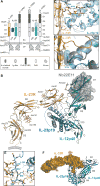
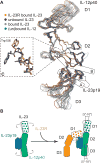
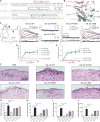
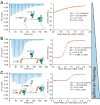
Interleukin-23 (IL-23), an IL-12 family cytokine, plays pivotal roles in pro-inflammatory T helper 17 cell responses linked to autoimmune and inflammatory diseases. Despite intense therapeutic targeting, structural and mechanistic insights into receptor complexes mediated by IL-23, and by IL-12 family members in general, have remained elusive. We determined a crystal structure of human IL-23 in complex with its cognate receptor, IL-23R, and revealed that IL-23R bound to IL-23 exclusively via its N-terminal immunoglobulin domain. The structural and functional hotspot of this interaction partially restructured the helical IL-23p19 subunit of IL-23 and restrained its IL-12p40 subunit to cooperatively bind the shared receptor IL-12Rβ1 with high affinity. Together with structural insights from the interaction of IL-23 with the inhibitory antibody briakinumab and by leveraging additional IL-23:antibody complexes, we propose a mechanistic paradigm for IL-23 and IL-12 whereby cognate receptor binding to the helical cytokine subunits primes recruitment of the shared receptors via the IL-12p40 subunit.
Keywords: Crohn’s disease; IL-12Rβ1; IL-23; IL-23 receptor; cytokine-receptor complex; inflammation; inflammatory bowel disease; psoriasis; rheumatoid arthritis; structure.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Deciphering site 3 interactions of interleukin 12 and interleukin 23 with their cognate murine and human receptors.J Biol Chem. 2020 Jul 24;295(30):10478-10492. doi: 10.1074/jbc.RA120.013935. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518162 Free PMC article.
-
Non-canonical interleukin 23 receptor complex assembly: p40 protein recruits interleukin 12 receptor β1 via site II and induces p19/interleukin 23 receptor interaction via site III.J Biol Chem. 2015 Jan 2;290(1):359-70. doi: 10.1074/jbc.M114.617597. Epub 2014 Nov 4. J Biol Chem. 2015. PMID: 25371211 Free PMC article.
-
Tryptophan (W) at position 37 of murine IL-12/IL-23 p40 is mandatory for binding to IL-12Rβ1 and subsequent signal transduction.J Biol Chem. 2021 Nov;297(5):101295. doi: 10.1016/j.jbc.2021.101295. Epub 2021 Oct 9. J Biol Chem. 2021. PMID: 34637790 Free PMC article.
-
Interleukin-23: a key cytokine in inflammatory diseases.Ann Med. 2011 Nov;43(7):503-11. doi: 10.3109/07853890.2011.577093. Epub 2011 May 17. Ann Med. 2011. PMID: 21585245 Review.
-
Regulation of Memory T Cells by Interleukin-23.Int Arch Allergy Immunol. 2016;169(3):157-62. doi: 10.1159/000445834. Epub 2016 Apr 22. Int Arch Allergy Immunol. 2016. PMID: 27100864 Review.
Cited by
-
Small-Format Drug Conjugates: A Viable Alternative to ADCs for Solid Tumours?Antibodies (Basel). 2018 Mar 31;7(2):16. doi: 10.3390/antib7020016. Antibodies (Basel). 2018. PMID: 31544868 Free PMC article. Review.
-
Psoriasis and Leprosy: An Arcane Relationship.J Inflamm Res. 2023 Jun 14;16:2521-2533. doi: 10.2147/JIR.S407650. eCollection 2023. J Inflamm Res. 2023. PMID: 37337513 Free PMC article.
-
Novel sampling strategies and a coarse-grained score function for docking homomers, flexible heteromers, and oligosaccharides using Rosetta in CAPRI rounds 37-45.Proteins. 2020 Aug;88(8):973-985. doi: 10.1002/prot.25855. Epub 2019 Dec 3. Proteins. 2020. PMID: 31742764 Free PMC article.
-
Deciphering site 3 interactions of interleukin 12 and interleukin 23 with their cognate murine and human receptors.J Biol Chem. 2020 Jul 24;295(30):10478-10492. doi: 10.1074/jbc.RA120.013935. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518162 Free PMC article.
-
Structural Basis for How Biologic Medicines Bind their Targets in Psoriasis Therapy.Yale J Biol Med. 2020 Mar 27;93(1):19-27. eCollection 2020 Mar. Yale J Biol Med. 2020. PMID: 32226331 Free PMC article.
References
-
- Arakawa A, Ruzicka T, Prinz JC. Therapeutic Efficacy of Interleukin 12/Interleukin 23 Blockade in Generalized Pustular Psoriasis Regardless of IL36RN Mutation Status. JAMA Dermatol. 2016;152:825–828. - PubMed
-
- Aricescu aR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta crystallographica. Section D, Biological crystallography. 2006;62:12431250. - PubMed
-
- Benson JM, Sachs CW, Treacy G, Zhou H, Pendley CE, Brodmerkel CM, Shankar G, Mascelli MA. Therapeutic targeting of the IL-12/23 pathways: generation and characterization of ustekinumab. Nat Biotechnol. 2011;29:615–624. - PubMed
-
- Beyer BM, Ingram R, Ramanathan L, Reichert P, Le HV, Madison V, Orth P. Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody. Journal of molecular biology. 2008;382:942–955. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

