Exploring the molecular complexity of Triatoma dimidiata sialome
- PMID: 29288089
- PMCID: PMC5803406
- DOI: 10.1016/j.jprot.2017.12.016
Exploring the molecular complexity of Triatoma dimidiata sialome
Abstract
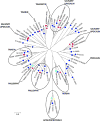
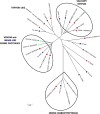
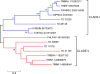
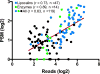
Triatoma dimidiata, a Chagas disease vector widely distributed along Central America, has great capability for domestic adaptation as the majority of specimens caught inside human dwellings or in peridomestic areas fed human blood. Exploring the salivary compounds that overcome host haemostatic and immune responses is of great scientific interest. Here, we provide a deeper insight into its salivary gland molecules. We used high-throughput RNA sequencing to examine in depth the T. dimidiata salivary gland transcriptome. From >51 million reads assembled, 92.21% are related to putative secreted proteins. Lipocalin is the most abundant gene family, confirming it is an expanded family in Triatoma genus salivary repertoire. Other putatively secreted members include phosphatases, odorant binding protein, hemolysin, proteases, protease inhibitors, antigen-5 and antimicrobial peptides. This work expands the previous set of functionally annotated sequences from T. dimidiata salivary glands available in NCBI from 388 to 3815. Additionally, we complemented the salivary analysis through proteomics (available data via ProteomeXchange with identifier PXD008510), disclosing the set complexity of 119 secreted proteins and validating the transcriptomic results. Our large-scale approach enriches the pharmacologically active molecules database and improves our knowledge about the complexity of salivary compounds from haematophagous vectors and their biological interactions.
Significance: Several haematophagous triatomine species can transmit Trypanosoma cruzi, the etiological agent of Chagas disease. Due to the reemergence of this disease, new drugs for its prevention and treatment are considered priorities. For this reason, the knowledge of vector saliva emerges as relevant biological finding, contributing to the design of different strategies for vector control and disease transmission. Here we report the transcriptomic and proteomic compositions of the salivary glands (sialome) of the reduviid bug Triatoma dimidiata, a relevant Chagas disease vector in Central America. Our results are robust and disclosed unprecedented insights into the notable diversity of its salivary glands content, revealing relevant anti-haemostatic salivary gene families. Our work expands almost ten times the previous set of functionally annotated sequences from T. dimidiata salivary glands available in NCBI. Moreover, using an integrated transcriptomic and proteomic approach, we showed a correlation pattern of transcription and translation processes for the main gene families found, an important contribution to the research of triatomine sialomes. Furthermore, data generated here reinforces the secreted proteins encountered can greatly contribute for haematophagic habit, Trypanosoma cruzi transmission and development of therapeutic agent studies.
Keywords: Chagas disease; Haematophagy; Sialome; Triatoma dimidiata; Triatominae; Vector biology.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures







References
-
- Bern C. Chagas’ Disease. N Engl J Med. 2015;373(5):456–66. - PubMed
-
- Dorn PL, Monroy C, Curtis A. Triatoma dimidiata (Latreille, 1811): a review of its diversity across its geographic range and the relationship among populations. Infect Genet Evol. 2007;7(2):343–52. - PubMed
-
- Ramírez CJ, et al. Genetic structure of sylvatic, peridomestic and domestic populations of Triatoma dimidiata (Hemiptera: Reduviidae) from an endemic zone of Boyaca. Colombia Acta Trop. 2005;93(1):23–9. - PubMed
-
- Guhl F. Chagas disease in Andean countries. Mem Inst Oswaldo Cruz. 2007;102(Suppl 1):29–38. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

