Rates of gut microbiome divergence in mammals
- PMID: 29290090
- PMCID: PMC5935551
- DOI: 10.1111/mec.14473
Rates of gut microbiome divergence in mammals
Abstract
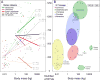
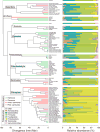
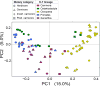
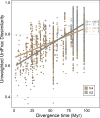
The variation and taxonomic diversity among mammalian gut microbiomes raises several questions about the factors that contribute to the rates and patterns of change in these microbial communities. By comparing the microbiome compositions of 112 species representing 14 mammalian orders, we assessed how host and ecological factors contribute to microbiome diversification. Except in rare cases, the same bacterial phyla predominate in mammalian gut microbiomes, and there has been some convergence of microbiome compositions according to dietary category across all mammalians lineages except Chiropterans (bats), which possess high proportions of Proteobacteria and tend to be most similar to one another regardless of diet. At lower taxonomic ranks (families, genera, 97% OTUs), bacteria are more likely to be associated with a particular mammalian lineage than with a particular dietary category, resulting in a strong phylogenetic signal in the degree to which microbiomes diverge. Despite different physiologies, the gut microbiomes of several mammalian lineages have diverged at roughly the same rate over the past 75 million years; however, the gut microbiomes of Cetartiodactyla (ruminants, whales, hippopotami) have evolved much faster and those of Chiropterans much slower. Contrary to expectations, the number of dietary transitions within a lineage does not influence rates of microbiome divergence, but instead, some of the most dramatic changes are associated with the loss of bacterial taxa, such as those accompanying the transition from terrestrial to marine lifestyles and the evolution of hominids.
Keywords: evolutionary rates; microbiome; molecular evolution; phylosymbiosis.
© 2017 John Wiley & Sons Ltd.
Figures

References
-
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Garber PA. The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta pigra) American Journal of Physical Anthropology. 2014;155:652–664. - PubMed
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 Available from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

