Stabilization of Reversed Replication Forks by Telomerase Drives Telomere Catastrophe
- PMID: 29290468
- PMCID: PMC5786504
- DOI: 10.1016/j.cell.2017.11.047
Stabilization of Reversed Replication Forks by Telomerase Drives Telomere Catastrophe
Abstract
Telomere maintenance critically depends on the distinct activities of telomerase, which adds telomeric repeats to solve the end replication problem, and RTEL1, which dismantles DNA secondary structures at telomeres to facilitate replisome progression. Here, we establish that reversed replication forks are a pathological substrate for telomerase and the source of telomere catastrophe in Rtel1-/- cells. Inhibiting telomerase recruitment to telomeres, but not its activity, or blocking replication fork reversal through PARP1 inhibition or depleting UBC13 or ZRANB3 prevents the rapid accumulation of dysfunctional telomeres in RTEL1-deficient cells. In this context, we establish that telomerase binding to reversed replication forks inhibits telomere replication, which can be mimicked by preventing replication fork restart through depletion of RECQ1 or PARG. Our results lead us to propose that telomerase inappropriately binds to and inhibits restart of reversed replication forks within telomeres, which compromises replication and leads to critically short telomeres.
Keywords: Hoyeraal-Hreidarsson syndrome; PARP1; RECQ1; RTEL1; ZRANB3; genome stability; replication fork reversal; telomerase; telomeres.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






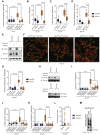
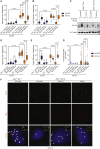
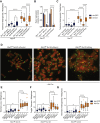
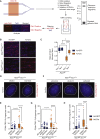
Comment in
-
When Telomerase Causes Telomere Loss.Dev Cell. 2018 Feb 5;44(3):281-283. doi: 10.1016/j.devcel.2018.01.011. Epub 2018 Feb 5. Dev Cell. 2018. PMID: 29408234
Similar articles
-
Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition.Nat Struct Mol Biol. 2013 Mar;20(3):347-54. doi: 10.1038/nsmb.2501. Epub 2013 Feb 10. Nat Struct Mol Biol. 2013. PMID: 23396353 Free PMC article.
-
Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability.Hum Mol Genet. 2013 Aug 15;22(16):3239-49. doi: 10.1093/hmg/ddt178. Epub 2013 Apr 15. Hum Mol Genet. 2013. PMID: 23591994
-
Full length RTEL1 is required for the elongation of the single-stranded telomeric overhang by telomerase.Nucleic Acids Res. 2020 Jul 27;48(13):7239-7251. doi: 10.1093/nar/gkaa503. Nucleic Acids Res. 2020. PMID: 32542379 Free PMC article.
-
RecQ Family Helicases in Replication Fork Remodeling and Repair: Opening New Avenues towards the Identification of Potential Targets for Cancer Chemotherapy.Anticancer Agents Med Chem. 2020;20(11):1311-1326. doi: 10.2174/1871520620666200518082433. Anticancer Agents Med Chem. 2020. PMID: 32418530 Review.
-
Telomere Replication: Solving Multiple End Replication Problems.Front Cell Dev Biol. 2021 Apr 1;9:668171. doi: 10.3389/fcell.2021.668171. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869233 Free PMC article. Review.
Cited by
-
Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases.Nucleic Acids Res. 2020 Jan 10;48(1):16-35. doi: 10.1093/nar/gkz1101. Nucleic Acids Res. 2020. PMID: 31754720 Free PMC article. Review.
-
Telomerase subunit Est2 marks internal sites that are prone to accumulate DNA damage.BMC Biol. 2021 Nov 20;19(1):247. doi: 10.1186/s12915-021-01167-1. BMC Biol. 2021. PMID: 34801008 Free PMC article.
-
Mechanisms and regulation of replication fork reversal.DNA Repair (Amst). 2024 Sep;141:103731. doi: 10.1016/j.dnarep.2024.103731. Epub 2024 Jul 22. DNA Repair (Amst). 2024. PMID: 39089193 Review.
-
How DNA damage and non-canonical nucleotides alter the telomerase catalytic cycle.DNA Repair (Amst). 2021 Nov;107:103198. doi: 10.1016/j.dnarep.2021.103198. Epub 2021 Jul 31. DNA Repair (Amst). 2021. PMID: 34371388 Free PMC article. Review.
-
Telomerase misbehaves after a breakup.Science. 2024 Feb 16;383(6684):702-703. doi: 10.1126/science.adn7791. Epub 2024 Feb 15. Science. 2024. PMID: 38359118 Free PMC article.
References
-
- Asai A., Oshima Y., Yamamoto Y., Uochi T.A., Kusaka H., Akinaga S., Yamashita Y., Pongracz K., Pruzan R., Wunder E. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res. 2003;63:3931–3939. - PubMed
-
- Ballew B.J., Joseph V., De S., Sarek G., Vannier J.B., Stracker T., Schrader K.A., Small T.N., O’Reilly R., Manschreck C. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet. 2013;9:e1003695. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

