Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor
- PMID: 29290469
- PMCID: PMC5766378
- DOI: 10.1016/j.cell.2017.12.004
Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor
Abstract
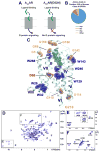
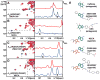
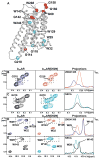
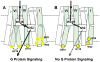
Signaling across cellular membranes, the 826 human G protein-coupled receptors (GPCRs) govern a wide range of vital physiological processes, making GPCRs prominent drug targets. X-ray crystallography provided GPCR molecular architectures, which also revealed the need for additional structural dynamics data to support drug development. Here, nuclear magnetic resonance (NMR) spectroscopy with the wild-type-like A2A adenosine receptor (A2AAR) in solution provides a comprehensive characterization of signaling-related structural dynamics. All six tryptophan indole and eight glycine backbone 15N-1H NMR signals in A2AAR were individually assigned. These NMR probes provided insight into the role of Asp522.50 as an allosteric link between the orthosteric drug binding site and the intracellular signaling surface, revealing strong interactions with the toggle switch Trp 2466.48, and delineated the structural response to variable efficacy of bound drugs across A2AAR. The present data support GPCR signaling based on dynamic interactions between two semi-independent subdomains connected by an allosteric switch at Asp522.50.
Keywords: G protein-coupled receptor; GPCR; NMR; allosteric modulation; membrane protein; nuclear magnetic resonance; signaling.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Affinity Mass Spectrometry-Based Fragment Screening Identified a New Negative Allosteric Modulator of the Adenosine A2A Receptor Targeting the Sodium Ion Pocket.ACS Chem Biol. 2021 Jun 18;16(6):991-1002. doi: 10.1021/acschembio.0c00899. Epub 2021 May 28. ACS Chem Biol. 2021. PMID: 34048655
-
Activation of adenosine A2A receptor by lipids from docosahexaenoic acid revealed by NMR.Sci Adv. 2020 Mar 18;6(12):eaay8544. doi: 10.1126/sciadv.aay8544. eCollection 2020 Mar. Sci Adv. 2020. PMID: 32206717 Free PMC article.
-
Extrinsic Tryptophans as NMR Probes of Allosteric Coupling in Membrane Proteins: Application to the A2A Adenosine Receptor.J Am Chem Soc. 2018 Jul 5;140(26):8228-8235. doi: 10.1021/jacs.8b03805. Epub 2018 Jun 20. J Am Chem Soc. 2018. PMID: 29874058 Free PMC article.
-
Novel approaches for targeting the adenosine A2A receptor.Expert Opin Drug Discov. 2015 Jan;10(1):63-80. doi: 10.1517/17460441.2015.971006. Epub 2014 Oct 14. Expert Opin Drug Discov. 2015. PMID: 25311639 Review.
-
Adenosine A2A receptor as a drug discovery target.J Med Chem. 2014 May 8;57(9):3623-50. doi: 10.1021/jm4011669. Epub 2013 Nov 15. J Med Chem. 2014. PMID: 24164628 Review.
Cited by
-
Delineating the conformational landscape of the adenosine A2A receptor during G protein coupling.Cell. 2021 Apr 1;184(7):1884-1894.e14. doi: 10.1016/j.cell.2021.02.041. Epub 2021 Mar 19. Cell. 2021. PMID: 33743210 Free PMC article.
-
The Conformational Equilibrium of the Neuropeptide Y2 Receptor in Bilayer Membranes.Angew Chem Int Ed Engl. 2020 Dec 21;59(52):23854-23861. doi: 10.1002/anie.202006075. Epub 2020 Sep 30. Angew Chem Int Ed Engl. 2020. PMID: 32790043 Free PMC article.
-
Capturing Peptide-GPCR Interactions and Their Dynamics.Molecules. 2020 Oct 15;25(20):4724. doi: 10.3390/molecules25204724. Molecules. 2020. PMID: 33076289 Free PMC article. Review.
-
Energy Landscapes Reveal Agonist Control of G Protein-Coupled Receptor Activation via Microswitches.Biochemistry. 2020 Feb 25;59(7):880-891. doi: 10.1021/acs.biochem.9b00842. Epub 2020 Feb 7. Biochemistry. 2020. PMID: 31999436 Free PMC article.
-
Tools shaping drug discovery and development.Biophys Rev (Melville). 2022 Jul 27;3(3):031301. doi: 10.1063/5.0087583. eCollection 2022 Sep. Biophys Rev (Melville). 2022. PMID: 38505278 Free PMC article. Review.
References
-
- Audet M, Bouvier M. Restructuring G-Protein-Coupled Receptor Activation. Cell. 2012;151:14–23. - PubMed
-
- Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosi. 1995;25:366–428.
-
- Bara-Jimenez W, Sherzai A, Dimitrova T, Favit A, Bibbiani F, Gillespie M, Morris M, Mouradian M, Chase T. Adenosine A2A receptor antagonist treatment of Parkinson’s disease. Neurology. 2003;61:293–296. - PubMed
-
- Beck M, Sakmar TP, Siebert F. Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry. 1998;37:7630–7639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

