CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis
- PMID: 29290589
- PMCID: PMC5791736
- DOI: 10.1016/j.devcel.2017.11.024
CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis
Abstract
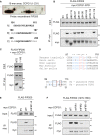
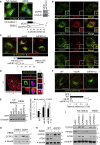
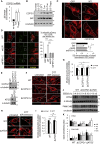
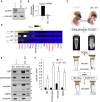
Mechanisms of selective autophagy of the ER, known as ER-phagy, require molecular delineation, particularly in vivo. It is unclear how these events control ER proteostasis and cellular health. Here, we identify cell-cycle progression gene 1 (CCPG1), an ER-resident protein with no known physiological role, as a non-canonical cargo receptor that directly binds to core autophagy proteins via an LIR motif to mammalian ATG8 proteins and, independently and via a discrete motif, to FIP200. These interactions facilitate ER-phagy. The CCPG1 gene is inducible by the unfolded protein response and thus directly links ER stress to ER-phagy. In vivo, CCPG1 protects against ER luminal protein aggregation and consequent unfolded protein response hyperactivation and tissue injury of the exocrine pancreas. Thus, via identification of this autophagy protein, we describe an unexpected molecular mechanism of ER-phagy and provide evidence that this may be physiologically relevant in ER luminal proteostasis.
Keywords: Atg8; CCPG1; ER-phagy; FIP200; autophagy; pancreas; proteostasis; tissue homeostasis; unfolded protein response.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
A Dual Binding Receptor for ER-phagy.Dev Cell. 2018 Jan 22;44(2):133-135. doi: 10.1016/j.devcel.2018.01.001. Dev Cell. 2018. PMID: 29401415
-
CCPG1 is a noncanonical autophagy cargo receptor essential for reticulophagy and pancreatic ER proteostasis.Autophagy. 2018;14(7):1107-1109. doi: 10.1080/15548627.2018.1467199. Autophagy. 2018. PMID: 29911925 Free PMC article.
References
-
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

