Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response
- PMID: 29290610
- PMCID: PMC5772982
- DOI: 10.1016/j.molcel.2017.12.002
Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response
Abstract
As sessile organisms, plants must adapt to variations in the environment. Environmental stress triggers various responses, including growth inhibition, mediated by the plant hormone abscisic acid (ABA). The mechanisms that integrate stress responses with growth are poorly understood. Here, we discovered that the Target of Rapamycin (TOR) kinase phosphorylates PYL ABA receptors at a conserved serine residue to prevent activation of the stress response in unstressed plants. This phosphorylation disrupts PYL association with ABA and with PP2C phosphatase effectors, leading to inactivation of SnRK2 kinases. Under stress, ABA-activated SnRK2s phosphorylate Raptor, a component of the TOR complex, triggering TOR complex dissociation and inhibition. Thus, TOR signaling represses ABA signaling and stress responses in unstressed conditions, whereas ABA signaling represses TOR signaling and growth during times of stress. Plants utilize this conserved phospho-regulatory feedback mechanism to optimize the balance of growth and stress responses.
Keywords: ABA receptor; Raptor; SnRK2; Target of Rapamycin; abscisic acid; phosphorylation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


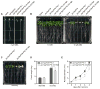

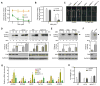
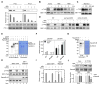

Comment in
-
To Grow or Not to Grow: TOR and SnRK2 Coordinate Growth and Stress Response in Arabidopsis.Mol Cell. 2018 Jan 4;69(1):3-4. doi: 10.1016/j.molcel.2017.12.013. Mol Cell. 2018. PMID: 29304332
References
-
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. - PubMed
-
- Antoni R, Rodriguez L, Gonzalez-Guzman M, Pizzio GA, Rodriguez PL. News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr Opin Plant Biol. 2011;14:547–553. - PubMed
-
- Assmann SM, Jegla T. Guard cell sensory systems: recent insights on stomatal responses to light, abscisic acid, and CO2. Curr Opin Plant Biol. 2016;33:157–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

