Meta-Analysis of MicroRNAs Dysregulated in the Hippocampal Dentate Gyrus of Animal Models of Epilepsy
- PMID: 29291240
- PMCID: PMC5745610
- DOI: 10.1523/ENEURO.0152-17.2017
Meta-Analysis of MicroRNAs Dysregulated in the Hippocampal Dentate Gyrus of Animal Models of Epilepsy
Abstract
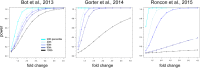
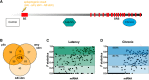
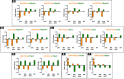
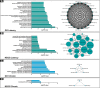
The identification of mechanisms transforming normal to seizure-generating tissue after brain injury is key to developing new antiepileptogenic treatments. MicroRNAs (miRNAs) may act as regulators and potential treatment targets for epileptogenesis. Here, we undertook a meta-analysis of changes in miRNA expression in the hippocampal dentate gyrus (DG) following an epileptogenic insult in three epilepsy models. We identified 26 miRNAs significantly differentially expressed during epileptogenesis, and five differentially expressed in chronic epilepsy. Of these, 13 were not identified in any of the individual studies. To assess the role of these miRNAs, we predicted their mRNA targets and then filtered the list to include only target genes expressed in DG and negatively correlated with miRNA expression. Functional enrichment analysis of mRNA targets of miRNAs dysregulated during epileptogenesis suggested a role for molecular processes related to inflammation and synaptic function. Our results identify new miRNAs associated with epileptogenesis from existing data, highlighting the utility of meta-analysis in maximizing value from preclinical data.
Keywords: dentate gyrus; epilepsy; hippocampus; mRNA; meta-analysis; miRNA.
Figures

Similar articles
-
Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy.Neurobiol Dis. 2014 Feb;62:508-20. doi: 10.1016/j.nbd.2013.10.026. Epub 2013 Oct 31. Neurobiol Dis. 2014. PMID: 24184920
-
MicroRNA profiling in the dentate gyrus in epileptic rats: The role of miR-187-3p.Medicine (Baltimore). 2017 Jun;96(22):e6744. doi: 10.1097/MD.0000000000006744. Medicine (Baltimore). 2017. PMID: 28562528 Free PMC article.
-
MicroRNAs and target genes in epileptogenesis.Epilepsia. 2020 Oct;61(10):2086-2096. doi: 10.1111/epi.16687. Epub 2020 Sep 18. Epilepsia. 2020. PMID: 32944964 Review.
-
MicroRNAs in epilepsy: pathophysiology and clinical utility.Lancet Neurol. 2016 Dec;15(13):1368-1376. doi: 10.1016/S1474-4422(16)30246-0. Lancet Neurol. 2016. PMID: 27839653 Review.
-
Integrative network analysis of miRNA-mRNA expression profiles during epileptogenesis in rats reveals therapeutic targets after emergence of first spontaneous seizure.Sci Rep. 2024 Jul 3;14(1):15313. doi: 10.1038/s41598-024-66117-7. Sci Rep. 2024. PMID: 38961125 Free PMC article.
Cited by
-
MicroRNA-335-5p suppresses voltage-gated sodium channel expression and may be a target for seizure control.Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2216658120. doi: 10.1073/pnas.2216658120. Epub 2023 Jul 18. Proc Natl Acad Sci U S A. 2023. PMID: 37463203 Free PMC article.
-
An Insight into the microRNAs Associated with Arteriovenous and Cavernous Malformations of the Brain.Cells. 2021 Jun 2;10(6):1373. doi: 10.3390/cells10061373. Cells. 2021. PMID: 34199498 Free PMC article. Review.
-
Genetic deletion of microRNA-22 blunts the inflammatory transcriptional response to status epilepticus and exacerbates epilepsy in mice.Mol Brain. 2020 Aug 21;13(1):114. doi: 10.1186/s13041-020-00653-x. Mol Brain. 2020. PMID: 32825833 Free PMC article.
-
Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps.Epilepsia Open. 2018 Oct 30;3(4):427-436. doi: 10.1002/epi4.12275. eCollection 2018 Dec. Epilepsia Open. 2018. PMID: 30525113 Free PMC article. Review.
-
Age-Related microRNA Overexpression in Lafora Disease Male Mice Provides Links between Neuroinflammation and Oxidative Stress.Int J Mol Sci. 2023 Jan 6;24(2):1089. doi: 10.3390/ijms24021089. Int J Mol Sci. 2023. PMID: 36674605 Free PMC article.
References
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
