Binding, folding and insertion of a β-hairpin peptide at a lipid bilayer surface: Influence of electrostatics and lipid tail packing
- PMID: 29291379
- PMCID: PMC5780206
- DOI: 10.1016/j.bbamem.2017.12.019
Binding, folding and insertion of a β-hairpin peptide at a lipid bilayer surface: Influence of electrostatics and lipid tail packing
Abstract
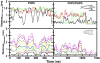
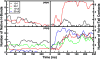
Antimicrobial peptides (AMPs) act as host defenses against microbial pathogens. Here we investigate the interactions of SVS-1 (KVKVKVKVdPlPTKVKVKVK), an engineered AMP and anti-cancer β-hairpin peptide, with lipid bilayers using spectroscopic studies and atomistic molecular dynamics simulations. In agreement with literature reports, simulation and experiment show preferential binding of SVS-1 peptides to anionic over neutral bilayers. Fluorescence and circular dichroism studies of a Trp-substituted SVS-1 analogue indicate, however, that it will bind to a zwitterionic DPPC bilayer under high-curvature conditions and folds into a hairpin. In bilayers formed from a 1:1 mixture of DPPC and anionic DPPG lipids, curvature and lipid fluidity are also observed to promote deeper insertion of the fluorescent peptide. Simulations using the CHARMM C36m force field offer complementary insight into timescales and mechanisms of folding and insertion. SVS-1 simulated at an anionic mixed POPC/POPG bilayer folded into a hairpin over a microsecond, the final stage in folding coinciding with the establishment of contact between the peptide's valine sidechains and the lipid tails through a "flip and dip" mechanism. Partial, transient folding and superficial bilayer contact are seen in simulation of the peptide at a zwitterionic POPC bilayer. Only when external surface tension is applied does the peptide establish lasting contact with the POPC bilayer. Our findings reveal the influence of disruption to lipid headgroup packing (via curvature or surface tension) on the pathway of binding and insertion, highlighting the collaborative effort of electrostatic and hydrophobic interactions on interaction of SVS-1 with lipid bilayers.
Keywords: Curvature; Fluorescence spectroscopy; Lipid bilayers; Liposome; SVS-1; Surface tension.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures







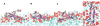
References
-
- Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacological Reviews. 2003;55(1):27–55. - PubMed
-
- Bond PJ, Khalid S. Antimicrobial and Cell-Penetrating Peptides: Structure, Assembly and Mechanisms of Membrane Lysis via Atomistic and Coarse-Grained Molecular Dynamic Simulations. Protein and Peptide Letters. 2010;17(11):1313–1327. - PubMed
-
- Giuliani A, Pirri G, Nicoletto S. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent Eur J Biol. 2007;2(1):1–33.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

