Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies
- PMID: 29292991
- PMCID: PMC5985209
- DOI: 10.1021/acs.chemrev.6b00750
Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies
Abstract
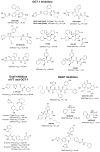
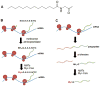
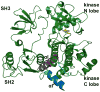
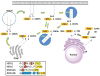
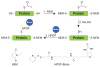
Protein lipidation, including cysteine prenylation, N-terminal glycine myristoylation, cysteine palmitoylation, and serine and lysine fatty acylation, occurs in many proteins in eukaryotic cells and regulates numerous biological pathways, such as membrane trafficking, protein secretion, signal transduction, and apoptosis. We provide a comprehensive review of protein lipidation, including descriptions of proteins known to be modified and the functions of the modifications, the enzymes that control them, and the tools and technologies developed to study them. We also highlight key questions about protein lipidation that remain to be answered, the challenges associated with answering such questions, and possible solutions to overcome these challenges.
Figures














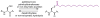


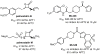
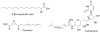






References
-
- Lentz BR. Exposure of Platelet Membrane Phosphatidylserine Regulates Blood Coagulation. Prog Lipid Res. 2003;42:423–438. - PubMed
-
- Menon AK, Kinoshita T, Orlean PA, Tamanoi F. Glycosylphosphatidylinositol (Gpi) Anchoring of Proteins. Academic Press; 2009.
-
- Zurzolo C, Simons K. Glycosylphosphatidylinositol-Anchored Proteins: Membrane Organization and Transport. Biochim Biophys Acta. 2016;1858:632–639. - PubMed
-
- Paladino S, Lebreton S, Zurzolo C. Trafficking and Membrane Organization of Gpi-Anchored Proteins in Health and Diseases. Curr Top Membr. 2015;75:269–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

