High throughput SNP discovery and genotyping in hexaploid wheat
- PMID: 29293495
- PMCID: PMC5749704
- DOI: 10.1371/journal.pone.0186329
High throughput SNP discovery and genotyping in hexaploid wheat
Abstract
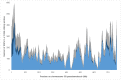
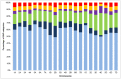
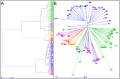
Because of their abundance and their amenability to high-throughput genotyping techniques, Single Nucleotide Polymorphisms (SNPs) are powerful tools for efficient genetics and genomics studies, including characterization of genetic resources, genome-wide association studies and genomic selection. In wheat, most of the previous SNP discovery initiatives targeted the coding fraction, leaving almost 98% of the wheat genome largely unexploited. Here we report on the use of whole-genome resequencing data from eight wheat lines to mine for SNPs in the genic, the repetitive and non-repetitive intergenic fractions of the wheat genome. Eventually, we identified 3.3 million SNPs, 49% being located on the B-genome, 41% on the A-genome and 10% on the D-genome. We also describe the development of the TaBW280K high-throughput genotyping array containing 280,226 SNPs. Performance of this chip was examined by genotyping a set of 96 wheat accessions representing the worldwide diversity. Sixty-nine percent of the SNPs can be efficiently scored, half of them showing a diploid-like clustering. The TaBW280K was proven to be a very efficient tool for diversity analyses, as well as for breeding as it can discriminate between closely related elite varieties. Finally, the TaBW280K array was used to genotype a population derived from a cross between Chinese Spring and Renan, leading to the construction a dense genetic map comprising 83,721 markers. The results described here will provide the wheat community with powerful tools for both basic and applied research.
Conflict of interest statement
Figures
References
-
- Ganal MW, Altmann T, Röder MS. SNP identification in crop plants. Curr Opin Plant Biol. 2009;12: 211–217. doi: 10.1016/j.pbi.2008.12.009 - DOI - PubMed
-
- Ganal MW, Polley A, Graner EM, Plieske J, Wieseke R, Luerssen H, et al. Large SNP arrays for genotyping in crop plants. J Biosci. 2012;37: 821–828. - PubMed
-
- Paux E, Sourdille P, Mackay I, Feuillet C. Sequence-based marker development in wheat: advances and applications to breeding. Biotechnol Adv. 2011;30: 1071–1088. doi: 10.1016/j.biotechadv.2011.09.015 - DOI - PubMed
-
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43: 956–963. doi: 10.1038/ng.911 - DOI - PubMed
-
- Kim N, Jeong YM, Jeong S, Kim GB, Baek S, Kwon YE, et al. Identification of candidate domestication regions in the radish genome based on high-depth resequencing analysis of 17 genotypes. Theor Appl Genet. 2016;129: 1797–1814. doi: 10.1007/s00122-016-2741-z - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

