PARP10 (ARTD10) modulates mitochondrial function
- PMID: 29293500
- PMCID: PMC5749700
- DOI: 10.1371/journal.pone.0187789
PARP10 (ARTD10) modulates mitochondrial function
Abstract
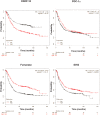
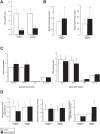
Poly(ADP-ribose) polymerase (PARP)10 is a PARP family member that performs mono-ADP-ribosylation of target proteins. Recent studies have linked PARP10 to metabolic processes and metabolic regulators that prompted us to assess whether PARP10 influences mitochondrial oxidative metabolism. The depletion of PARP10 by specific shRNAs increased mitochondrial oxidative capacity in cellular models of breast, cervical, colorectal and exocrine pancreas cancer. Upon silencing of PARP10, mitochondrial superoxide production decreased in line with increased expression of antioxidant genes pointing out lower oxidative stress upon PARP10 silencing. Improved mitochondrial oxidative capacity coincided with increased AMPK activation. The silencing of PARP10 in MCF7 and CaCo2 cells decreased the proliferation rate that correlated with increased expression of anti-Warburg enzymes (Foxo1, PGC-1α, IDH2 and fumarase). By analyzing an online database we showed that lower PARP10 expression increases survival in gastric cancer. Furthermore, PARP10 expression decreased upon fasting, a condition that is characterized by increases in mitochondrial biogenesis. Finally, lower PARP10 expression is associated with increased fatty acid oxidation.
Conflict of interest statement
Figures
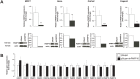
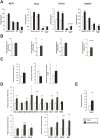

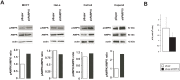
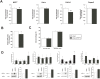

References
-
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–19 doi: 10.1016/j.tibs.2009.12.003 - DOI - PubMed
-
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32(1):57–69 doi: 10.1016/j.molcel.2008.08.009 - DOI - PubMed
-
- Bai P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol Cell. 2015;58(6):947–58 doi: 10.1016/j.molcel.2015.01.034 - DOI - PubMed
-
- Virag L, Salzman AL, Szabo C. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. JImmunol. 1998;161(7):3753–9 - PubMed
-
- Bai P, Nagy L, Fodor T, Liaudet L, Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab. 2015;26(2):75–83 doi: 10.1016/j.tem.2014.11.003 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

