Prospects for Genomic Selection in Cassava Breeding
- PMID: 29293806
- PMCID: PMC7822052
- DOI: 10.3835/plantgenome2017.03.0015
Prospects for Genomic Selection in Cassava Breeding
Abstract
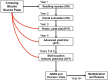
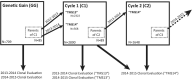
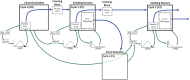
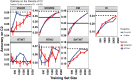
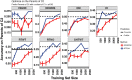
Cassava ( Crantz) is a clonally propagated staple food crop in the tropics. Genomic selection (GS) has been implemented at three breeding institutions in Africa to reduce cycle times. Initial studies provided promising estimates of predictive abilities. Here, we expand on previous analyses by assessing the accuracy of seven prediction models for seven traits in three prediction scenarios: cross-validation within populations, cross-population prediction and cross-generation prediction. We also evaluated the impact of increasing the training population (TP) size by phenotyping progenies selected either at random or with a genetic algorithm. Cross-validation results were mostly consistent across programs, with nonadditive models predicting of 10% better on average. Cross-population accuracy was generally low (mean = 0.18) but prediction of cassava mosaic disease increased up to 57% in one Nigerian population when data from another related population were combined. Accuracy across generations was poorer than within-generation accuracy, as expected, but accuracy for dry matter content and mosaic disease severity should be sufficient for rapid-cycling GS. Selection of a prediction model made some difference across generations, but increasing TP size was more important. With a genetic algorithm, selection of one-third of progeny could achieve an accuracy equivalent to phenotyping all progeny. We are in the early stages of GS for this crop but the results are promising for some traits. General guidelines that are emerging are that TPs need to continue to grow but phenotyping can be done on a cleverly selected subset of individuals, reducing the overall phenotyping burden.
Copyright © 2017 Crop Science Society of America.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


References
-
- Akano O., Dixon O, Mba C, Barrera E, and Fregene M. 2002. Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 105(4):521–525. doi:10.1007/s00122-002-0891-7 - DOI - PubMed
-
- Akdemir D., and Okeke U.G. 2015. EMMREML : Fitting mixed models with known covariance structures. https://CRAN.R-project.org/package=EMMREML (accessed 30 Aug. 2017).
-
- Akdemir D., Sanchez J.I, and Jannink J.-L. 2015. Optimization of genomic selection training populations with a genetic algorithm. Genet. Sel. Evol. 47(1):38. doi:10.1186/s12711-015-0116-6 - DOI - PMC - PubMed
-
- Bamidele O.P., Fasogbon M.B, Oladiran D.A, and Akande E.O. 2015. Nutritional composition of fufu analog flour produced from Cassava root (Manihot esculenta) and Cocoyam (Colocasia esculenta) tuber. Food Sci. Nutr. 3(6):597–603. doi:10.1002/fsn3.250 - DOI - PMC - PubMed
-
- Barabaschi D., Tondelli A, Desiderio F, Volante A, Vaccino P, Vale G, et al. . 2015. Next generation breeding. Plant Sci. 242:3–13. doi:10.1016/j.plantsci.2015.07.010 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

