An Evolutionary Landscape of A-to-I RNA Editome across Metazoan Species
- PMID: 29294013
- PMCID: PMC5800060
- DOI: 10.1093/gbe/evx277
An Evolutionary Landscape of A-to-I RNA Editome across Metazoan Species
Abstract
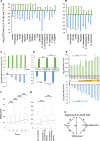
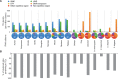
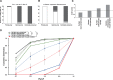
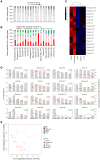
Adenosine-to-inosine (A-to-I) editing is widespread across the kingdom Metazoa. However, for the lack of comprehensive analysis in nonmodel animals, the evolutionary history of A-to-I editing remains largely unexplored. Here, we detect high-confidence editing sites using clustering and conservation strategies based on RNA sequencing data alone, without using single-nucleotide polymorphism information or genome sequencing data from the same sample. We thereby unveil the first evolutionary landscape of A-to-I editing maps across 20 metazoan species (from worm to human), providing unprecedented evidence on how the editing mechanism gradually expands its territory and increases its influence along the history of evolution. Our result revealed that highly clustered and conserved editing sites tended to have a higher editing level and a higher magnitude of the ADAR motif. The ratio of the frequencies of nonsynonymous editing to that of synonymous editing remarkably increased with increasing the conservation level of A-to-I editing. These results thus suggest potentially functional benefit of highly clustered and conserved editing sites. In addition, spatiotemporal dynamics analyses reveal a conserved enrichment of editing and ADAR expression in the central nervous system throughout more than 300 Myr of divergent evolution in complex animals and the comparability of editing patterns between invertebrates and between vertebrates during development. This study provides evolutionary and dynamic aspects of A-to-I editome across metazoan species, expanding this important but understudied class of nongenomically encoded events for comprehensive characterization.
Keywords: A-to-I RNA editing; ADAR; ADAR motif; dynamic editome; evolution.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Bahn JH, et al. 2012. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22(1):142–150.http://dx.doi.org/10.1101/gr.124107.111 - DOI - PMC - PubMed
-
- Bass B, et al. 2012. The difficult calls in RNA editing. Interviewed by H Craig Mak. Nat Biotechnol. 30(12):1207–1209.http://dx.doi.org/10.1038/nbt.2452 - DOI - PubMed
-
- Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 71:817–846.http://dx.doi.org/10.1146/annurev.biochem.71.110601.135501 - DOI - PMC - PubMed
-
- Bazak L, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24:365–376.http://dx.doi.org/10.1101/gr.164749.113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

