Integrative analysis reveals disrupted pathways regulated by microRNAs in cancer
- PMID: 29294105
- PMCID: PMC5814839
- DOI: 10.1093/nar/gkx1250
Integrative analysis reveals disrupted pathways regulated by microRNAs in cancer
Abstract
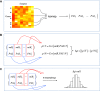
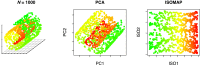
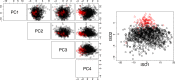
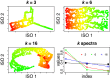
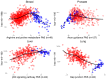
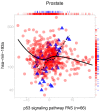
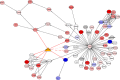
MicroRNAs (miRNAs) are small endogenous regulatory molecules that modulate gene expression post-transcriptionally. Although differential expression of miRNAs have been implicated in many diseases (including cancers), the underlying mechanisms of action remain unclear. Because each miRNA can target multiple genes, miRNAs may potentially have functional implications for the overall behavior of entire pathways. Here, we investigate the functional consequences of miRNA dysregulation through an integrative analysis of miRNA and mRNA expression data using a novel approach that incorporates pathway information a priori. By searching for miRNA-pathway associations that differ between healthy and tumor tissue, we identify specific relationships at the systems level which are disrupted in cancer. Our approach is motivated by the hypothesis that if an miRNA and pathway are associated, then the expression of the miRNA and the collective behavior of the genes in a pathway will be correlated. As such, we first obtain an expression-based summary of pathway activity using Isomap, a dimension reduction method which can articulate non-linear structure in high-dimensional data. We then search for miRNAs that exhibit differential correlations with the pathway summary between phenotypes as a means of finding aberrant miRNA-pathway coregulation in tumors. We apply our method to cancer data using gene and miRNA expression datasets from The Cancer Genome Atlas and compare ∼105 miRNA-pathway relationships between healthy and tumor samples from four tissues (breast, prostate, lung and liver). Many of the flagged pairs we identify have a biological basis for disruption in cancer.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

