Carbon Catabolite Repression in Filamentous Fungi
- PMID: 29295552
- PMCID: PMC5795998
- DOI: 10.3390/ijms19010048
Carbon Catabolite Repression in Filamentous Fungi
Abstract
Carbon Catabolite Repression (CCR) has fascinated scientists and researchers around the globe for the past few decades. This important mechanism allows preferential utilization of an energy-efficient and readily available carbon source over relatively less easily accessible carbon sources. This mechanism helps microorganisms to obtain maximum amount of glucose in order to keep pace with their metabolism. Microorganisms assimilate glucose and highly favorable sugars before switching to less-favored sources of carbon such as organic acids and alcohols. In CCR of filamentous fungi, CreA acts as a transcription factor, which is regulated to some extent by ubiquitination. CreD-HulA ubiquitination ligase complex helps in CreA ubiquitination, while CreB-CreC deubiquitination (DUB) complex removes ubiquitin from CreA, which causes its activation. CCR of fungi also involves some very crucial elements such as Hexokinases, cAMP, Protein Kinase (PKA), Ras proteins, G protein-coupled receptor (GPCR), Adenylate cyclase, RcoA and SnfA. Thorough study of molecular mechanism of CCR is important for understanding growth, conidiation, virulence and survival of filamentous fungi. This review is a comprehensive revision of the regulation of CCR in filamentous fungi as well as an updated summary of key regulators, regulation of different CCR-dependent mechanisms and its impact on various physical characteristics of filamentous fungi.
Keywords: CreA; cAMP; carbon catabolite repression; hexokinase; phosphorylation; sensing and signaling pathway; transport proteins; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest. The authors declare that the research is being conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
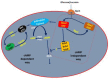
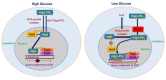
 represents protein interaction in the nucleus, mig1 and hxk2, Ssn6-Tup1 co-repressor complex and
represents protein interaction in the nucleus, mig1 and hxk2, Ssn6-Tup1 co-repressor complex and  mig1. Shows that there is no protein interaction, mig1 and hxk2, Ssn6-Tup1 co-repressor complex and mig1 and between Ssn6-Tup1 co-repressor complex and promoter gene.
mig1. Shows that there is no protein interaction, mig1 and hxk2, Ssn6-Tup1 co-repressor complex and mig1 and between Ssn6-Tup1 co-repressor complex and promoter gene.  represents the inhibition of transcription [34,45,46,47,48]. During high glucose conditions, inactive Snf1 cannot cause phosphorylation of Mig1 and cellular movement of Mig1 is dependent on Glc7–Reg1 complex; whereas during low glucose levels Snf1 will be active and cause direct repression of Snf1, which will be unable to repress the CCR subjected genes.
represents the inhibition of transcription [34,45,46,47,48]. During high glucose conditions, inactive Snf1 cannot cause phosphorylation of Mig1 and cellular movement of Mig1 is dependent on Glc7–Reg1 complex; whereas during low glucose levels Snf1 will be active and cause direct repression of Snf1, which will be unable to repress the CCR subjected genes.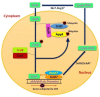
References
-
- Matar K.A.O., Chen X., Chen D., Anjago W.M., Norvienyeku J., Lin Y., Chen M., Wang Z., Ebbole D.J., Lu G. WD40-repeat protein MoCreC is essential for carbon repression and is involved in conidiation, growth and pathogenicity of Magnaporthe oryzae. Curr. Genet. 2017;63:685–696. doi: 10.1007/s00294-016-0668-1. - DOI - PubMed
-
- Portnoy T., Margeot A., Linke R., Atanasova L., Fekete E., Sándor E., Hartl L., Karaffa L., Druzhinina I.S., Seiboth B. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: A master regulator of carbon assimilation. BMC Genom. 2011;12:269. doi: 10.1186/1471-2164-12-269. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

