Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats
- PMID: 29295979
- PMCID: PMC5750222
- DOI: 10.1038/s41467-017-02441-z
Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats
Abstract
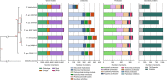
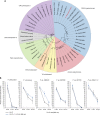
Bat white-nose syndrome (WNS), caused by the fungal pathogen Pseudogymnoascus destructans, has decimated North American hibernating bats since its emergence in 2006. Here, we utilize comparative genomics to examine the evolutionary history of this pathogen in comparison to six closely related nonpathogenic species. P. destructans displays a large reduction in carbohydrate-utilizing enzymes (CAZymes) and in the predicted secretome (~50%), and an increase in lineage-specific genes. The pathogen has lost a key enzyme, UVE1, in the alternate excision repair (AER) pathway, which is known to contribute to repair of DNA lesions induced by ultraviolet (UV) light. Consistent with a nonfunctional AER pathway, P. destructans is extremely sensitive to UV light, as well as the DNA alkylating agent methyl methanesulfonate (MMS). The differential susceptibility of P. destructans to UV light in comparison to other hibernacula-inhabiting fungi represents a potential "Achilles' heel" of P. destructans that might be exploited for treatment of bats with WNS.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon. 2009;108:147–154. doi: 10.5248/108.147. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

