Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis
- PMID: 29296188
- PMCID: PMC5746090
- DOI: 10.18632/oncotarget.22534
Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis
Abstract
Background: Fungi colonize the human gut and might play a key role in the pathogenesis of ulcerative colitis (UC). However, studies on the fungal composition in the gut (especially adhering to the intestinal mucosa) of UC patients is limited.
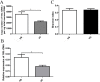
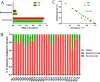
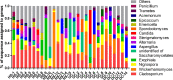
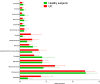
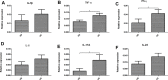
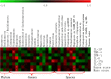
Results: The number of fungi decreased significantly in inflamed mucosa compared with that in HS mucosa. Fifteen major genera were examined, among which Wickerhamomyces, unidentified genus of Saccharomycetales, Aspergillus, Sterigmatomyces, and Candida showed increasing trends, whereas Exophiala, Alternaria, Emericella, Epicoccum, Acremonium, Trametes, and Penicillium showed decreasing trends in UC patients compared to the HS. The pro-inflammatory cytokines (IL-Iβ, TNF-α, INF-γ, IL-6, IL-17A, and IL-23) were up-regulated in the UC group. The genera Wickerhamomyces, Nigrospora, and Penicillium were positively correlated, while Sporobolomyces and Trametes were negatively correlated with the expression of several colonic pro-inflammatory cytokines and the Baron and/or Mayo score.
Conclusions: Our study confirms the alteration of the colonic fungal microbiota in the UC patients, which might be associated with mucosal inflammation and pathogenesis of UC. Further studies need to identify the roles of different intestinal fungi in detail, and to determine the mechanism of the host-fungal interaction underlying the development of UC.
Methods: Mucosal samples of inflamed descending colon from 14 active UC patients and 15 healthy subjects (HS) were analyzed by high-throughput sequencing to compare the fungal microbiota. The expressions of pro-inflammatory cytokines (IL-Iβ, TNF-α, INF-γ, IL-6, IL-17A, and IL-23) in intestinal mucosal tissues were examined. The Baron and Mayo scores of UC patients were evaluated, and the correlation between intestinal fungal composition and intestinal inflammatory status was analyzed.
Keywords: high-throughput sequencing; intestinal fungi; mucosal inflammation; mucosal-associated microbiota; ulcerative colitis.
Conflict of interest statement
CONFLICTS OF INTEREST None.
Figures

References
-
- Qiu X, Ma J, Wang K, Zhang H. Chemopreventive effects of 5-aminosalicylic acid on inflammatory bowel disease-associated colorectal cancer and dysplasia: a systematic review with meta-analysis. Oncotarget. 2017;8:1031–1045. http://doi.org/10.18632/oncotarget.13715. - DOI - PMC - PubMed
-
- Casellas F, Lopez-Vivancos J, Casado A, Malagelada JR. Factors affecting health related quality of life of patients with inflammatory bowel disease. Qual Life Res. 2002;11:775–781. - PubMed
-
- Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C, Niewiadomski O, Bell S, Kamm MA, de Silva HJ, Kasturiratne A, Senanayake YU, Ooi CJ, et al. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut. 2015;64:1063–1071. - PubMed
-
- Cammarota G, Ianiro G, Cianci R, Bibbo S, Gasbarrini A, Curro D. The involvement of gut microbiota in inflammatory bowel disease pathogenesis: potential for therapy. Pharmacol Ther. 2015;149:191–212. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

