Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project
- PMID: 29296939
- PMCID: PMC5737168
- DOI: 10.1182/bloodadvances.2016001867
Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project
Abstract
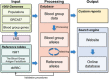
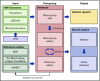
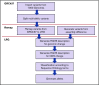
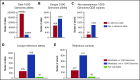
Blood group genotyping has recently developed into a clinical tool to improve compatibility of blood transfusions and management of pregnancies. Next-generation sequencing (NGS) is rapidly moving toward routine practice for patient and donor typing and has the potential to remedy some of the limitations of currently used platforms. However, a large-scale investigation into the blood group genotypes obtained by NGS in a multiethnic cohort is lacking. The 1000 Genomes Project provides information on genome variation among 2504 individuals representing 26 populations worldwide. We extracted their NGS data for all 36 blood group systems to a custom-designed database. In total, 210 412 alleles from 43 blood group-related genes were imported and curated. Matching algorithms were developed to compare them to blood group variants identified to date. Of the 1241 non-synonymous variants identified in the coding regions, 241 are known blood group polymorphisms. Interestingly, 357 of the remaining 1000 variants are predicted to occur on extracellular portions of 31 different blood group-carrying proteins and some may represent undiscovered antigens. Of the alleles analyzed, 1504 were not previously described. The ABO/GBGT1/FUT2/FUT3 and GYPB/GYPC genes showed the highest degree of variation per kilobase coding sequence, and ACKR1 variants had the most skewed distribution across 5 continental superpopulations in the dataset. Results were exported to an online search engine, www.erythrogene.com, which presents data according to the allele nomenclature developed for clinical reporting by the International Society of Blood Transfusion. The established database deepens our knowledge on blood group polymorphism globally and provides a long-sought platform for future research.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
References
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931-945. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

