A database of human genes and a gene network involved in response to tick-borne encephalitis virus infection
- PMID: 29297316
- PMCID: PMC5751789
- DOI: 10.1186/s12862-017-1107-8
A database of human genes and a gene network involved in response to tick-borne encephalitis virus infection
Abstract
Background: Tick-borne encephalitis is caused by the neurotropic, positive-sense RNA virus, tick-borne encephalitis virus (TBEV). TBEV infection can lead to a variety of clinical manifestations ranging from slight fever to severe neurological illness. Very little is known about genetic factors predisposing to severe forms of disease caused by TBEV. The aims of the study were to compile a catalog of human genes involved in response to TBEV infection and to rank genes from the catalog based on the number of neighbors in the network of pairwise interactions involving these genes and TBEV RNA or proteins.
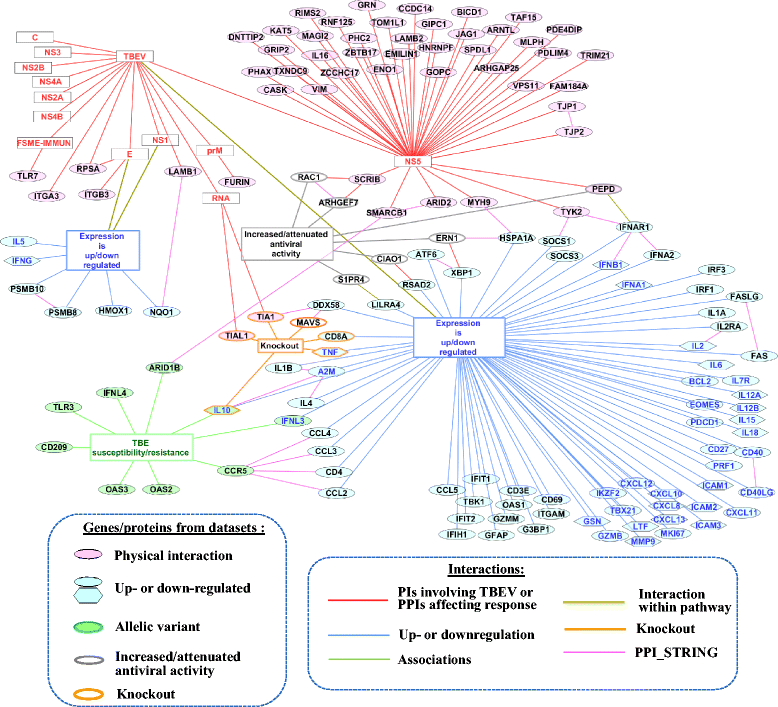
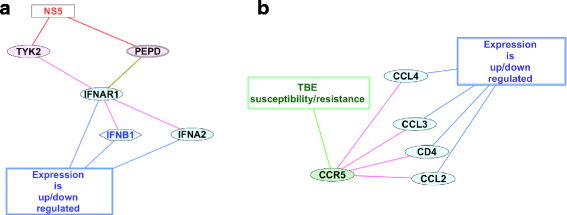
Results: Based on manual review and curation of scientific publications a catalog comprising 140 human genes involved in response to TBEV infection was developed. To provide access to data on all genes, the TBEVhostDB web resource ( http://icg.nsc.ru/TBEVHostDB/ ) was created. We reconstructed a network formed by pairwise interactions between TBEV virion itself, viral RNA and viral proteins and 140 genes/proteins from TBEVHostDB. Genes were ranked according to the number of interactions in the network. Two genes/proteins (CCR5 and IFNAR1) that had maximal number of interactions were revealed. It was found that the subnetworks formed by CCR5 and IFNAR1 and their neighbors were a fragments of two key pathways functioning during the course of tick-borne encephalitis: (1) the attenuation of interferon-I signaling pathway by the TBEV NS5 protein that targeted peptidase D; (2) proinflammation and tissue damage pathway triggered by chemokine receptor CCR5 interacting with CD4, CCL3, CCL4, CCL2. Among nine genes associated with severe forms of TBEV infection, three genes/proteins (CCR5, IL10, ARID1B) were found to have protein-protein interactions within the network, and two genes/proteins (IFNL3 and the IL10, that was just mentioned) were up- or down-regulated in response to TBEV infection. Based on this finding, potential mechanisms for participation of CCR5, IL10, ARID1B, and IFNL3 in the host response to TBEV infection were suggested.
Conclusions: A database comprising 140 human genes involved in response to TBEV infection was compiled and the TBEVHostDB web resource, providing access to all genes was created. This is the first effort of integrating and unifying data on genetic factors that may predispose to severe forms of diseases caused by TBEV. The TBEVHostDB could potentially be used for assessment of risk factors for severe forms of tick-borne encephalitis and for the design of personalized pharmacological strategies for the treatment of TBEV infection.
Keywords: Candidate genes; Database; Flavivirus; Network; PPIs; TBEV; Tick-borne encephalitis.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

