Enhanced Solubilization of Class B Radical S-Adenosylmethionine Methylases by Improved Cobalamin Uptake in Escherichia coli
- PMID: 29298049
- PMCID: PMC5941297
- DOI: 10.1021/acs.biochem.7b01205
Enhanced Solubilization of Class B Radical S-Adenosylmethionine Methylases by Improved Cobalamin Uptake in Escherichia coli
Abstract
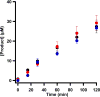
The methylation of unactivated carbon and phosphorus centers is a burgeoning area of biological chemistry, especially given that such reactions constitute key steps in the biosynthesis of numerous enzyme cofactors, antibiotics, and other natural products of clinical value. These kinetically challenging reactions are catalyzed exclusively by enzymes in the radical S-adenosylmethionine (SAM) superfamily and have been grouped into four classes (A-D). Class B radical SAM (RS) methylases require a cobalamin cofactor in addition to the [4Fe-4S] cluster that is characteristic of RS enzymes. However, their poor solubility upon overexpression and their generally poor turnover has hampered detailed in vitro studies of these enzymes. It has been suggested that improper folding, possibly caused by insufficient cobalamin during their overproduction in Escherichia coli, leads to formation of inclusion bodies. Herein, we report our efforts to improve the overproduction of class B RS methylases in a soluble form by engineering a strain of E. coli to take in more cobalamin. We cloned five genes ( btuC, btuE, btuD, btuF, and btuB) that encode proteins that are responsible for cobalamin uptake and transport in E. coli and co-expressed these genes with those that encode TsrM, Fom3, PhpK, and ThnK, four class B RS methylases that suffer from poor solubility during overproduction. This strategy markedly enhances the uptake of cobalamin into the cytoplasm and improves the solubility of the target enzymes significantly.
Figures







References
-
- Gomez Macqueo Chew A, Bryant DA. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Ann Rev Microbiol. 2007;61:113–129. - PubMed
-
- Kudo F, Kasama Y, Hirayama T, Eguchi T. Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J Antibiot. 2007;60:492–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

