CRISPRed Macrophages for Cell-Based Cancer Immunotherapy
- PMID: 29298051
- PMCID: PMC6063311
- DOI: 10.1021/acs.bioconjchem.7b00768
CRISPRed Macrophages for Cell-Based Cancer Immunotherapy
Abstract
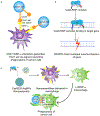
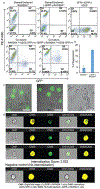
We present here an integrated nanotechnology/biology strategy for cancer immunotherapy that uses arginine nanoparticles (ArgNPs) to deliver CRISPR-Cas9 gene editing machinery into cells to generate SIRP-α knockout macrophages. The NP system efficiently codelivers single guide RNA (sgRNA) and Cas9 protein required for editing to knock out the "don't eat me signal" in macrophages that prevents phagocytosis of cancer cells. Turning off this signal increased the innate phagocytic capabilities of the macrophages by 4-fold. This improved attack and elimination of cancer cells makes this strategy promising for the creation of "weaponized" macrophages for cancer immunotherapy.
Figures

Similar articles
-
[Advances in application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 system in stem cells research].Zhonghua Shao Shang Za Zhi. 2018 Apr 20;34(4):253-256. doi: 10.3760/cma.j.issn.1009-2587.2018.04.013. Zhonghua Shao Shang Za Zhi. 2018. PMID: 29690746 Review. Chinese.
-
LeishGEdit: A Method for Rapid Gene Knockout and Tagging Using CRISPR-Cas9.Methods Mol Biol. 2019;1971:189-210. doi: 10.1007/978-1-4939-9210-2_9. Methods Mol Biol. 2019. PMID: 30980304
-
Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9.Protein Cell. 2017 Sep;8(9):634-643. doi: 10.1007/s13238-017-0410-x. Epub 2017 Apr 22. Protein Cell. 2017. PMID: 28434148 Free PMC article. Review.
-
Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):12514-12519. doi: 10.1073/pnas.1613884113. Epub 2016 Oct 11. Proc Natl Acad Sci U S A. 2016. PMID: 27729526 Free PMC article.
-
Recent advances in CRISPR/Cas9 mediated genome editing in Bacillus subtilis.World J Microbiol Biotechnol. 2018 Sep 29;34(10):153. doi: 10.1007/s11274-018-2537-1. World J Microbiol Biotechnol. 2018. PMID: 30269229 Review.
Cited by
-
Polarization of macrophages to an anti-cancer phenotype through in situ uncaging of a TLR 7/8 agonist using bioorthogonal nanozymes.Chem Sci. 2024 Jan 9;15(7):2486-2494. doi: 10.1039/d3sc06431j. eCollection 2024 Feb 14. Chem Sci. 2024. PMID: 38362405 Free PMC article.
-
Macrophage activation by a substituted pyrimido[5,4-b]indole increases anti-cancer activity.Pharmacol Res. 2019 Oct;148:104452. doi: 10.1016/j.phrs.2019.104452. Epub 2019 Sep 10. Pharmacol Res. 2019. PMID: 31518642 Free PMC article.
-
CAR-macrophage versus CAR-T for solid tumors: The race between a rising star and a superstar.Biomol Biomed. 2024 May 2;24(3):465-476. doi: 10.17305/bb.2023.9675. Biomol Biomed. 2024. PMID: 37877819 Free PMC article. Review.
-
Bioengineered therapeutic systems for improving antitumor immunity.Natl Sci Rev. 2024 Nov 12;12(1):nwae404. doi: 10.1093/nsr/nwae404. eCollection 2025 Jan. Natl Sci Rev. 2024. PMID: 40114728 Free PMC article. Review.
-
Delivering the CRISPR/Cas9 system for engineering gene therapies: Recent cargo and delivery approaches for clinical translation.Front Bioeng Biotechnol. 2022 Sep 26;10:973326. doi: 10.3389/fbioe.2022.973326. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36225598 Free PMC article. Review.
References
-
- Willingham SB, Volkmer J-P, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD et al. (2012) The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. U.S.A 109, 6662–6667. - PMC - PubMed
-
- Fehervari Z (2015) Don’t eat me, activate me. Nat. Immunol 16, 1113–1113.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

