G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs?
- PMID: 29298813
- PMCID: PMC5820538
- DOI: 10.1124/mol.117.111062
G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs?
Abstract
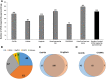
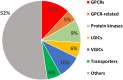
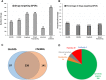
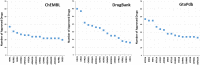
Estimates vary regarding the number of G protein-coupled receptors (GPCRs), the largest family of membrane receptors that are targeted by approved drugs, and the number of such drugs that target GPCRs. We review current knowledge regarding GPCRs as drug targets by integrating data from public databases (ChEMBL, Guide to PHARMACOLOGY, and DrugBank) and from the Broad Institute Drug Repurposing Hub. To account for discrepancies among these sources, we curated a list of GPCRs currently targeted by approved drugs. As of November 2017, 134 GPCRs are targets for drugs approved in the United States or European Union; 128 GPCRs are targets for drugs listed in the Food and Drug Administration Orange Book. We estimate that ∼700 approved drugs target GPCRs, implying that approximately 35% of approved drugs target GPCRs. GPCRs and GPCR-related proteins, i.e., those upstream of or downstream from GPCRs, represent ∼17% of all protein targets for approved drugs, with GPCRs themselves accounting for ∼12%. As such, GPCRs constitute the largest family of proteins targeted by approved drugs. Drugs that currently target GPCRs and GPCR-related proteins are primarily small molecules and peptides. Since ∼100 of the ∼360 human endo-GPCRs (other than olfactory, taste, and visual GPCRs) are orphan receptors (lacking known physiologic agonists), the number of GPCR targets, the number of GPCR-targeted drugs, and perhaps the types of drugs will likely increase, thus further expanding this GPCR repertoire and the many roles of GPCR drugs in therapeutics.
Copyright © 2018 by The American Society for Pharmacology and Experimental Therapeutics.
Figures
References
-
- Allen JA, Roth BL. (2011) Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol 51:117–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

