At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a Conserved Missing Link in the Regulation of Floral Meristem Termination in Arabidopsis and Tomato
- PMID: 29298836
- PMCID: PMC5810569
- DOI: 10.1105/tpc.17.00653
At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a Conserved Missing Link in the Regulation of Floral Meristem Termination in Arabidopsis and Tomato
Abstract
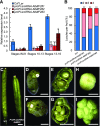
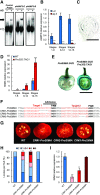
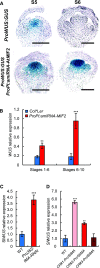
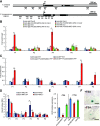
In angiosperms, the gynoecium is the last structure to develop within the flower due to the determinate fate of floral meristem (FM) stem cells. The maintenance of stem cell activity before its arrest at the stage called FM termination affects the number of carpels that develop. The necessary inhibition at this stage of WUSCHEL (WUS), which is responsible for stem cell maintenance, involves a two-step mechanism. Direct repression mediated by the MADS domain transcription factor AGAMOUS (AG), followed by indirect repression requiring the C2H2 zinc-finger protein KNUCKLES (KNU), allow for the complete termination of floral stem cell activity. Here, we show that Arabidopsis thaliana MINI ZINC FINGER2 (AtMIF2) and its homolog in tomato (Solanum lycopersicum), INHIBITOR OF MERISTEM ACTIVITY (SlIMA), participate in the FM termination process by functioning as adaptor proteins. AtMIF2 and SlIMA recruit AtKNU and SlKNU, respectively, to form a transcriptional repressor complex together with TOPLESS and HISTONE DEACETYLASE19. AtMIF2 and SlIMA bind to the WUS and SlWUS loci in the respective plants, leading to their repression. These results provide important insights into the molecular mechanisms governing (FM) termination and highlight the essential role of AtMIF2/SlIMA during this developmental step, which determines carpel number and therefore fruit size.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures






Comment in
-
A Conserved Mechanism to Terminate Floral Meristems.Plant Cell. 2018 Feb;30(2):260. doi: 10.1105/tpc.18.00054. Epub 2018 Jan 24. Plant Cell. 2018. PMID: 29367306 Free PMC article. No abstract available.
References
-
- Bereterbide A., Hernould M., Farbos I., Glimelius K., Mouras A. (2002). Restoration of stamen development and production of functional pollen in an alloplasmic CMS tobacco line by ectopic expression of the Arabidopsis thaliana SUPERMAN gene. Plant J. 29: 607–615. - PubMed
-
- Bisbis B., Delmas F., Joubès J., Sicard A., Hernould M., Inzé D., Mouras A., Chevalier C. (2006). Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J. Biol. Chem. 281: 7374–7383. - PubMed
-
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

