Linking invasive motility to protein expression in single tumor cells
- PMID: 29299576
- PMCID: PMC5771853
- DOI: 10.1039/c7lc01008g
Linking invasive motility to protein expression in single tumor cells
Abstract
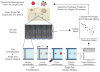
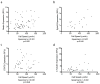
The invasion of malignant cells into tissue is a critical step in the progression of cancer. While it is increasingly appreciated that cells within a tumor differ in their invasive potential, it remains nearly unknown how these differences relate to cell-to-cell variations in protein expression. Here, we introduce a microfluidic platform that integrates measurements of invasive motility and protein expression for single cells, which we use to scrutinize human glioblastoma tumor-initiating cells (TICs). Our live-cell imaging microdevice is comprised of polyacrylamide microchannels that exhibit tissue-like stiffness and present chemokine gradients along each channel. Due to intrinsic differences in motility, cell subpopulations separate along the channel axis. The separated cells are then lysed in situ and each single-cell lysate is subjected to western blotting in the surrounding polyacrylamide matrix. We observe correlations between motility and Nestin and EphA2 expression. We identify protein-protein correlations within single TICs, which would be obscured with population-based assays. The integration of motility traits with single-cell protein analysis - on the same cell - offers a new means to identify druggable targets of invasive capacity.
Conflict of interest statement
There are no conflicts to declare
Figures
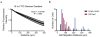

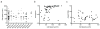


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

