The Gateway from Near into Remote Oceania: New Insights from Genome-Wide Data
- PMID: 29301001
- PMCID: PMC5889034
- DOI: 10.1093/molbev/msx333
The Gateway from Near into Remote Oceania: New Insights from Genome-Wide Data
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
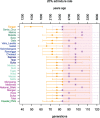
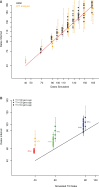
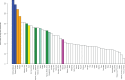
A widely accepted two-wave scenario of human settlement of Oceania involves the first out-of-Africa migration circa 50,000 years ago (ya), and the more recent Austronesian expansion, which reached the Bismarck Archipelago by 3,450 ya. Whereas earlier genetic studies provided evidence for extensive sex-biased admixture between the incoming and the indigenous populations, some archaeological, linguistic, and genetic evidence indicates a more complicated picture of settlement. To study regional variation in Oceania in more detail, we have compiled a genome-wide data set of 823 individuals from 72 populations (including 50 populations from Oceania) and over 620,000 autosomal single nucleotide polymorphisms (SNPs). We show that the initial dispersal of people from the Bismarck Archipelago into Remote Oceania occurred in a "leapfrog" fashion, completely by-passing the main chain of the Solomon Islands, and that the colonization of the Solomon Islands proceeded in a bidirectional manner. Our results also support a divergence between western and eastern Solomons, in agreement with the sharp linguistic divide known as the Tryon-Hackman line. We also report substantial post-Austronesian gene flow across the Solomons. In particular, Santa Cruz (in Remote Oceania) exhibits extraordinarily high levels of Papuan ancestry that cannot be explained by a simple bottleneck/founder event scenario. Finally, we use simulations to show that discrepancies between different methods for dating admixture likely reflect different sensitivities of the methods to multiple admixture events from the same (or similar) sources. Overall, this study points to the importance of fine-scale sampling to understand the complexities of human population history.
Figures




References
-
- Bellwood P. 2004. First farmers: the origins of agricultural societies. 1st ed Malden (MA: ): Wiley-Blackwell.
-
- Bellwood P, Dizon EZ.. 2005. The Batanes archaeological project and the Out of Taiwan hypothesis for Austronesian dispersal. J Austronesian Stud 1:1–31.
-
- Blust R. 1999. Subgrouping, circularity and extinction: some issues in Austronesian comparative linguistics. Symp Ser Inst Linguist Acad Sin 1:31–94.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

