Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers
- PMID: 29301303
- PMCID: PMC5796072
- DOI: 10.3390/ijms19010123
Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers
Abstract
Natural antisense transcripts are RNA sequences that can be transcribed from both DNA strands at the same locus but in the opposite direction from the gene transcript. Because strand-specific high-throughput sequencing of the antisense transcriptome has only been available for less than a decade, many natural antisense transcripts were first described as long non-coding RNAs. Although the precise biological roles of natural antisense transcripts are not known yet, an increasing number of studies report their implication in gene expression regulation. Their expression levels are altered in many physiological and pathological conditions, including breast cancers. Among the potential clinical utilities of the natural antisense transcripts, the non-coding|coding transcript pairs are of high interest for treatment. Indeed, these pairs can be targeted by antisense oligonucleotides to specifically tune the expression of the coding-gene. Here, we describe the current knowledge about natural antisense transcripts, their varying molecular mechanisms as gene expression regulators, and their potential as prognostic or predictive biomarkers in breast cancers.
Keywords: breast cancer; gene expression regulation; lncRNA; natural antisense transcript; natural antisense transcripts; next generation sequencing; non-coding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


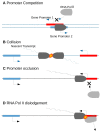

References
-
- ENCODE Project Consortium. Birney E., Stamatoyannopoulos J.A., Dutta A., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

