The comparison of alternative splicing among the multiple tissues in cucumber
- PMID: 29301488
- PMCID: PMC5755334
- DOI: 10.1186/s12870-017-1217-x
The comparison of alternative splicing among the multiple tissues in cucumber
Abstract
Background: Alternative splicing (AS) is an important post-transcriptional process. It has been suggested that most AS events are subject to tissue-specific regulation. However, the global dynamics of AS in different tissues are poorly explored.
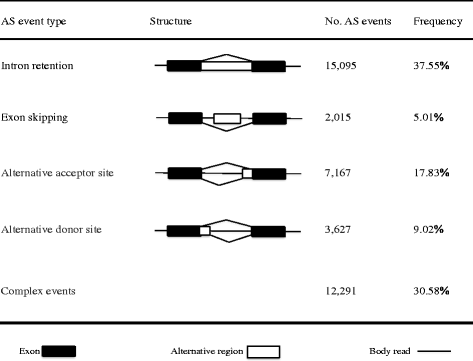
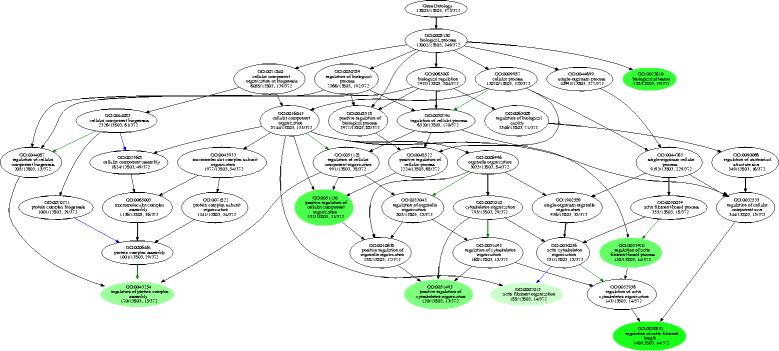
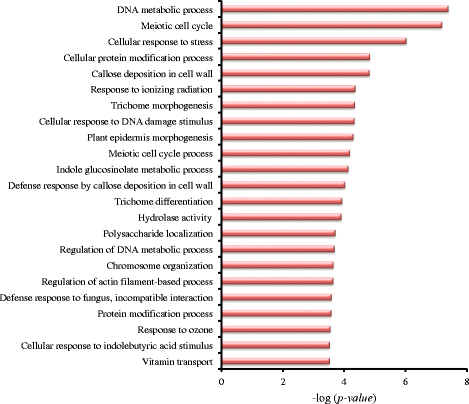
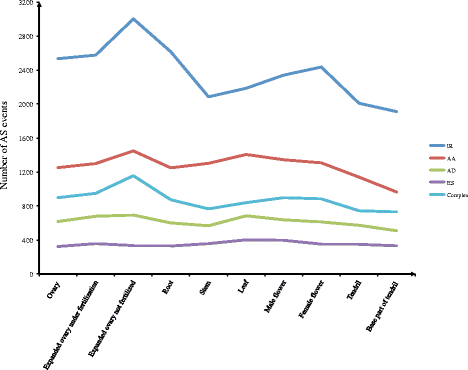
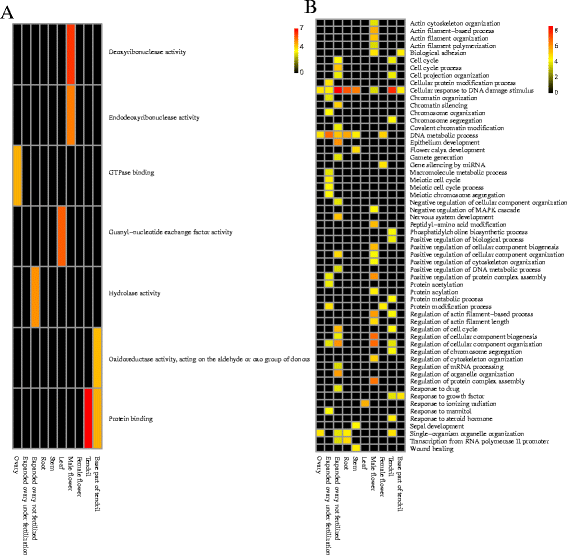
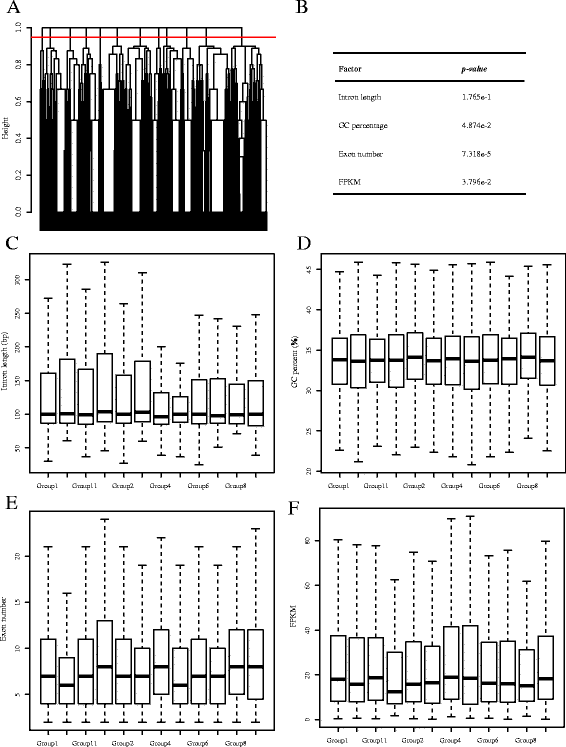
Results: To analyse global changes in AS in multiple tissues, we identified the AS events and constructed a comprehensive catalogue of AS events within each tissue based on the genome-wide RNA-seq reads from ten tissues in cucumber. First, we found that 58% of the multi-exon genes underwent AS. We further obtained 565 genes with significantly more AS events compared with random genes. These genes were found significant enrichment in biological processes related to the regulation of actin filament length. Second, significantly different AS event profiles among ten tissues were found. The tissues with the same origin of development are more likely to have a relatively similar AS profile. Moreover, 7370 genes showed tissue-specific AS events and were highly enriched in biological processes related to the positive regulation of cellular component organization. Root-specificity AS genes were related to the cellular response to DNA damage stimulus. Third, the genes with different intron retention (IR) patterns among the ten tissues showed significant difference in GC percentages of the retained intron, and the number of exons and FPKM of the major transcripts.
Conclusions: Our study provided a comprehensive view of AS in multiple tissues. We revealed novel insights into the patterns of AS in multiple tissues and the tissue-specific AS in cucumber.
Keywords: Alternative splicing; Cucumber; Tissue-specific; Tissues.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Prediction of transcript isoforms and identification of tissue-specific genes in cucumber.BMC Genomics. 2025 Jan 10;26(1):25. doi: 10.1186/s12864-025-11212-w. BMC Genomics. 2025. PMID: 39794760 Free PMC article.
-
Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus.PLoS One. 2012;7(10):e47576. doi: 10.1371/journal.pone.0047576. Epub 2012 Oct 23. PLoS One. 2012. PMID: 23110079 Free PMC article.
-
CuAS: a database of annotated transcripts generated by alternative splicing in cucumbers.BMC Plant Biol. 2020 Mar 18;20(1):119. doi: 10.1186/s12870-020-2312-y. BMC Plant Biol. 2020. PMID: 32183712 Free PMC article.
-
Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber.BMC Plant Biol. 2019 Aug 7;19(1):345. doi: 10.1186/s12870-019-1953-1. BMC Plant Biol. 2019. PMID: 31390991 Free PMC article.
-
The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation.J Pineal Res. 2014 Jan;56(1):39-50. doi: 10.1111/jpi.12095. Epub 2013 Oct 15. J Pineal Res. 2014. PMID: 24102657
Cited by
-
Single-molecule real-time sequencing facilitates the analysis of transcripts and splice isoforms of anthers in Chinese cabbage (Brassica rapa L. ssp. pekinensis).BMC Plant Biol. 2019 Nov 27;19(1):517. doi: 10.1186/s12870-019-2133-z. BMC Plant Biol. 2019. PMID: 31771515 Free PMC article.
-
Tissue-specific mouse mRNA isoform networks.Sci Rep. 2019 Sep 27;9(1):13949. doi: 10.1038/s41598-019-50119-x. Sci Rep. 2019. PMID: 31562339 Free PMC article.
-
A Global Analysis of Alternative Splicing of Dichocarpum Medicinal Plants, Ranunculales.Curr Genomics. 2022 Jul 5;23(3):207-216. doi: 10.2174/1389202923666220527112929. Curr Genomics. 2022. PMID: 36777007 Free PMC article.
-
Comprehensive transcriptomic and proteomic analyses identify intracellular targets for myriocin to induce Fusarium oxysporum f. sp. niveum cell death.Microb Cell Fact. 2021 Mar 17;20(1):69. doi: 10.1186/s12934-021-01560-z. Microb Cell Fact. 2021. PMID: 33731109 Free PMC article.
-
Prediction of transcript isoforms and identification of tissue-specific genes in cucumber.BMC Genomics. 2025 Jan 10;26(1):25. doi: 10.1186/s12864-025-11212-w. BMC Genomics. 2025. PMID: 39794760 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

