Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax
- PMID: 29302006
- PMCID: PMC5788258
- DOI: 10.1126/science.aan1078
Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax
Abstract
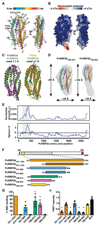
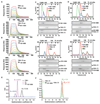
Plasmodium vivax shows a strict host tropism for reticulocytes. We identified transferrin receptor 1 (TfR1) as the receptor for P. vivax reticulocyte-binding protein 2b (PvRBP2b). We determined the structure of the N-terminal domain of PvRBP2b involved in red blood cell binding, elucidating the molecular basis for TfR1 recognition. We validated TfR1 as the biological target of PvRBP2b engagement by means of TfR1 expression knockdown analysis. TfR1 mutant cells deficient in PvRBP2b binding were refractory to invasion of P. vivax but not to invasion of P. falciparum Using Brazilian and Thai clinical isolates, we show that PvRBP2b monoclonal antibodies that inhibit reticulocyte binding also block P. vivax entry into reticulocytes. These data show that TfR1-PvRBP2b invasion pathway is critical for the recognition of reticulocytes during P. vivax invasion.
Copyright © 2018, American Association for the Advancement of Science.
Figures



References
-
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. - PubMed
-
- Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

