Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells
- PMID: 29302013
- PMCID: PMC5933048
- DOI: 10.1126/science.aam8825
Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells
Erratum in
-
Erratum for the Report: "Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells" by V. Ramsuran, V. Naranbhai, A. Horowitz, Y. Qi, M. P. Martin, Y. Yuki, X. Gao, V. Walker-Sperling, G. Q. Del Prete, D. K. Schneider, J. D. Lifson, J. Fellay, S. G. Deeks, J. N. Martin, J. J. Goedert, S. M. Wolinsky, N. L. Michael, G. D. Kirk, S. Buchbinder, D. Haas, T. Ndung'u, P. Goulder, P. Parham, B. D. Walker, J. M. Carlson, M. Carrington.Science. 2019 Aug 2;365(6452):eaay7985. doi: 10.1126/science.aay7985. Science. 2019. PMID: 31371579 No abstract available.
Abstract
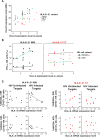
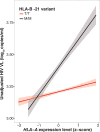
The highly polymorphic human leukocyte antigen (HLA) locus encodes cell surface proteins that are critical for immunity. HLA-A expression levels vary in an allele-dependent manner, diversifying allele-specific effects beyond peptide-binding preference. Analysis of 9763 HIV-infected individuals from 21 cohorts shows that higher HLA-A levels confer poorer control of HIV. Elevated HLA-A expression provides enhanced levels of an HLA-A-derived signal peptide that specifically binds and determines expression levels of HLA-E, the ligand for the inhibitory NKG2A natural killer (NK) cell receptor. HLA-B haplotypes that favor NKG2A-mediated NK cell licensing (i.e., education) exacerbate the deleterious effect of high HLA-A on HIV control, consistent with NKG2A-mediated inhibition impairing NK cell clearance of HIV-infected targets. Therapeutic blockade of HLA-E:NKG2A interaction may yield benefit in HIV disease.
Copyright © 2017, American Association for the Advancement of Science.
Figures
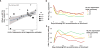
References
-
- Raj P, et al. eLife. 2016;5:e12089. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

