Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves
- PMID: 29303957
- PMCID: PMC5796095
- DOI: 10.3390/ijms19010146
Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves
Abstract
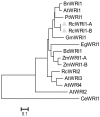
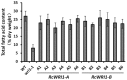
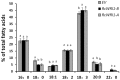
The plant-specific WRINKLED1 (WRI1) is a member of the AP2/EREBP class of transcription factors that positively regulate oil biosynthesis in plant tissues. Limited information is available for the role of WRI1 in oil biosynthesis in castor bean (Ricinus connunis L.), an important industrial oil crop. Here, we report the identification of two alternatively spliced transcripts of RcWRI1, designated as RcWRI1-A and RcWRI1-B. The open reading frames of RcWRI1-A (1341 bp) and RcWRI1-B (1332 bp) differ by a stretch of 9 bp, such that the predicted RcWRI1-B lacks the three amino acid residues "VYL" that are present in RcWRI1-A. The RcWRI1-A transcript is present in flowers, leaves, pericarps and developing seeds, while the RcWRI1-B mRNA is only detectable in developing seeds. When the two isoforms were individually introduced into an Arabidopsiswri1-1 loss-of-function mutant, total fatty acid content was almost restored to the wild-type level, and the percentage of the wrinkled seeds was largely reduced in the transgenic lines relative to the wri1-1 mutant line. Transient expression of each RcWRI1 splice isoform in N. benthamiana leaves upregulated the expression of the WRI1 target genes, and consequently increased the oil content by 4.3-4.9 fold when compared with the controls, and RcWRI1-B appeared to be more active than RcWRI1-A. Both RcWRI1-A and RcWRI1-B can be used as a key transcriptional regulator to enhance fatty acid and oil biosynthesis in leafy biomass.
Keywords: RcWRI1; alternative splice form; castor (Ricinus communis L.); fatty acid and oil biosynthesis; tobacco (Nicotiana benthamiana L.).
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Vanhercke T., El Tahchy A., Liu Q., Zhou X.R., Shrestha P., Divi U.K., Ral J.P., Mansour M.P., Nichols P.D., James C.N., et al. Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 2014;12:231–239. doi: 10.1111/pbi.12131. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

