Muscle-Specific Mis-Splicing and Heart Disease Exemplified by RBM20
- PMID: 29304022
- PMCID: PMC5793171
- DOI: 10.3390/genes9010018
Muscle-Specific Mis-Splicing and Heart Disease Exemplified by RBM20
Abstract
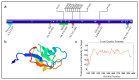
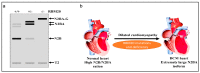
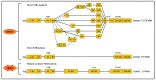
Alternative splicing is an essential post-transcriptional process to generate multiple functional RNAs or proteins from a single transcript. Progress in RNA biology has led to a better understanding of muscle-specific RNA splicing in heart disease. The recent discovery of the muscle-specific splicing factor RNA-binding motif 20 (RBM20) not only provided great insights into the general alternative splicing mechanism but also demonstrated molecular mechanism of how this splicing factor is associated with dilated cardiomyopathy. Here, we review our current knowledge of muscle-specific splicing factors and heart disease, with an emphasis on RBM20 and its targets, RBM20-dependent alternative splicing mechanism, RBM20 disease origin in induced Pluripotent Stem Cells (iPSCs), and RBM20 mutations in dilated cardiomyopathy. In the end, we will discuss the multifunctional role of RBM20 and manipulation of RBM20 as a potential therapeutic target for heart disease.
Keywords: RNA-binding motif 20; alternative splicing; heart disease; muscle-specific splicing factor; titin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

