Metabolic capability and in situ activity of microorganisms in an oil reservoir
- PMID: 29304850
- PMCID: PMC5756336
- DOI: 10.1186/s40168-017-0392-1
Metabolic capability and in situ activity of microorganisms in an oil reservoir
Abstract
Background: Microorganisms have long been associated with oxic and anoxic degradation of hydrocarbons in oil reservoirs and oil production facilities. While we can readily determine the abundance of microorganisms in the reservoir and study their activity in the laboratory, it has been challenging to resolve what microbes are actively participating in crude oil degradation in situ and to gain insight into what metabolic pathways they deploy.
Results: Here, we describe the metabolic potential and in situ activity of microbial communities obtained from the Jiangsu Oil Reservoir (China) by an integrated metagenomics and metatranscriptomics approach. Almost complete genome sequences obtained by differential binning highlight the distinct capability of different community members to degrade hydrocarbons under oxic or anoxic condition. Transcriptomic data delineate active members of the community and give insights that Acinetobacter species completely oxidize alkanes into carbon dioxide with the involvement of oxygen, and Archaeoglobus species mainly ferment alkanes to generate acetate which could be consumed by Methanosaeta species. Furthermore, nutritional requirements based on amino acid and vitamin auxotrophies suggest a complex network of interactions and dependencies among active community members that go beyond classical syntrophic exchanges; this network defines community composition and microbial ecology in oil reservoirs undergoing secondary recovery.
Conclusion: Our data expand current knowledge of the metabolic potential and role in hydrocarbon metabolism of individual members of thermophilic microbial communities from an oil reservoir. The study also reveals potential metabolic exchanges based on vitamin and amino acid auxotrophies indicating the presence of complex network of interactions between microbial taxa within the community.
Keywords: Auxotrophy; Hydrocarbon degradation; Metagenomics and metatranscriptomics; Microbial community; Oil reservoir.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
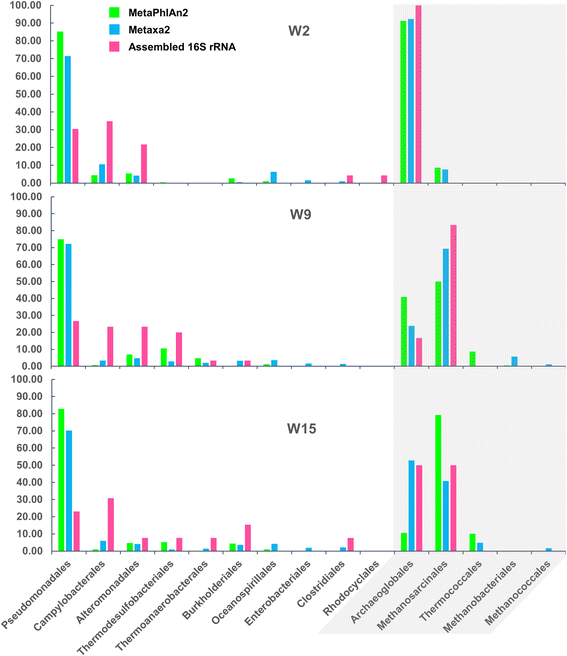

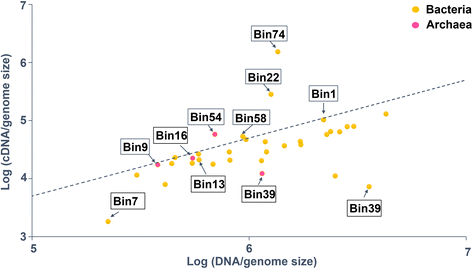
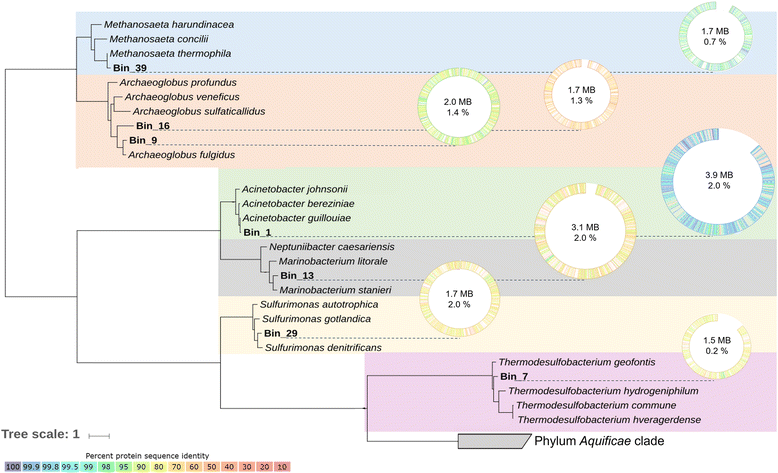
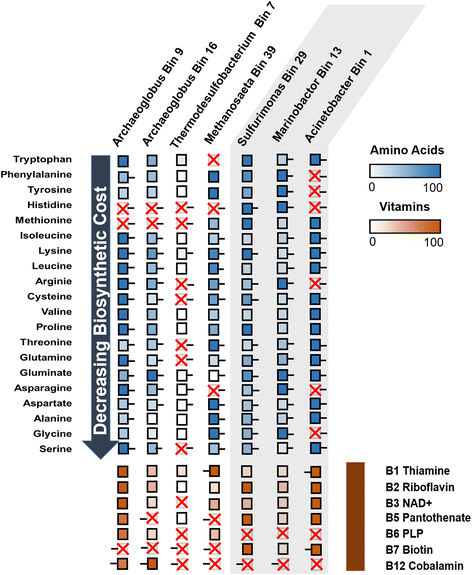
References
Publication types
MeSH terms
Substances
Grants and funding
- 41530318/National Natural Science Foundation of China/International
- 41373070/National Natural Science Foundation of China/International
- 41373070/National Natural Science Foundation of China/International
- 41161160560/NSFC/RGC Joint Research Fund/International
- 201011/2014-0/CNPq under the Brazilian Scientific Mobility Program, Ciências sem Fronteiras/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

