How Much Do rRNA Gene Surveys Underestimate Extant Bacterial Diversity?
- PMID: 29305502
- PMCID: PMC5835724
- DOI: 10.1128/AEM.00014-18
How Much Do rRNA Gene Surveys Underestimate Extant Bacterial Diversity?
Abstract
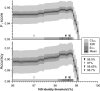
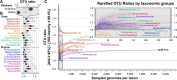
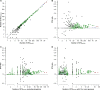
The most common practice in studying and cataloguing prokaryotic diversity involves the grouping of sequences into operational taxonomic units (OTUs) at the 97% 16S rRNA gene sequence identity level, often using partial gene sequences, such as PCR-generated amplicons. Due to the high sequence conservation of rRNA genes, organisms belonging to closely related yet distinct species may be grouped under the same OTU. However, it remains unclear how much diversity has been underestimated by this practice. To address this question, we compared the OTUs of genomes defined at the 97% or 98.5% 16S rRNA gene identity level against OTUs of the same genomes defined at the 95% whole-genome average nucleotide identity (ANI), which is a much more accurate proxy for species. Our results show that OTUs resulting from a 98.5% 16S rRNA gene identity cutoff are more accurate than 97% compared to 95% ANI (90.5% versus 89.9% accuracy) but indistinguishable from any other threshold in the 98.29 to 98.78% range. Even with the more stringent thresholds, however, the 16S rRNA gene-based approach commonly underestimates the number of OTUs by ∼12%, on average, compared to the ANI-based approach (∼14% underestimation when using the 97% identity threshold). More importantly, the degree of underestimation can become 50% or more for certain taxa, such as the genera Pseudomonas, Burkholderia, Escherichia, Campylobacter, and Citrobacter These results provide a quantitative view of the degree of underestimation of extant prokaryotic diversity by 16S rRNA gene-defined OTUs and suggest that genomic resolution is often necessary.IMPORTANCE Species diversity is one of the most fundamental pieces of information for community ecology and conservational biology. Therefore, employing accurate proxies for what a species or the unit of diversity is are cornerstones for a large set of microbial ecology and diversity studies. The most common proxies currently used rely on the clustering of 16S rRNA gene sequences at some threshold of nucleotide identity, typically 97% or 98.5%. Here, we explore how well this strategy reflects the more accurate whole-genome-based proxies and determine the frequency with which the high conservation of 16S rRNA sequences masks substantial species-level diversity.
Keywords: 16S rRNA gene; average nucleotide identity; diversity.
Copyright © 2018 American Society for Microbiology.
Figures


References
-
- Hubbell SP. 2008. The Unified neutral theory of biodiversity and biogeography (MPB-32). Princeton University Press, Princeton, NJ.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

