An IgaA/UmoB Family Protein from Serratia marcescens Regulates Motility, Capsular Polysaccharide Biosynthesis, and Secondary Metabolite Production
- PMID: 29305504
- PMCID: PMC5835751
- DOI: 10.1128/AEM.02575-17
An IgaA/UmoB Family Protein from Serratia marcescens Regulates Motility, Capsular Polysaccharide Biosynthesis, and Secondary Metabolite Production
Abstract
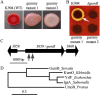
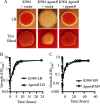
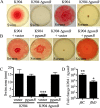
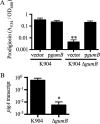
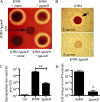
Secondary metabolites are an important source of pharmaceuticals and key modulators of microbe-microbe interactions. The bacterium Serratia marcescens is part of the Enterobacteriaceae family of eubacteria and produces a number of biologically active secondary metabolites. In this study, we screened for novel regulators of secondary metabolites synthesized by a clinical isolate of S. marcescens and found mutations in a gene for an uncharacterized UmoB/IgaA family member here named gumB Mutation of gumB conferred a severe loss of the secondary metabolites prodigiosin and serratamolide. The gumB mutation conferred pleiotropic phenotypes, including altered biofilm formation, highly increased capsular polysaccharide production, and loss of swimming and swarming motility. These phenotypes corresponded to transcriptional changes in fimA, wecA, and flhD Unlike other UmoB/IgaA family members, gumB was found to be not essential for growth in S. marcescens, yet igaA from Salmonella enterica, yrfF from Escherichia coli, and an uncharacterized predicted ortholog from Klebsiella pneumoniae complemented the gumB mutant secondary metabolite defects, suggesting highly conserved function. These data support the idea that UmoB/IgaA family proteins are functionally conserved and extend the known regulatory influence of UmoB/IgaA family proteins to the control of competition-associated secondary metabolites and biofilm formation.IMPORTANCE IgaA/UmoB family proteins are found in members of the Enterobacteriaceae family of bacteria, which are of environmental and public health importance. IgaA/UmoB family proteins are thought to be inner membrane proteins that report extracellular stresses to intracellular signaling pathways that respond to environmental challenge. This study introduces a new member of the IgaA/UmoB family and demonstrates a high degree of functional similarity between IgaA/UmoB family proteins. Moreover, this study extends the phenomena controlled by IgaA/UmoB family proteins to include the biosynthesis of antimicrobial secondary metabolites.
Keywords: biofilm; capsular polysaccharide; competition; flagella; motility; secondary metabolite; surfactant.
Copyright © 2018 American Society for Microbiology.
Figures


References
-
- Demain AL. 1992. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry. Ciba Found Symp 171:3–16, discussion 16–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

