HOTAIR Long Non-coding RNA: Characterizing the Locus Features by the In Silico Approaches
- PMID: 29307144
- PMCID: PMC5769859
- DOI: 10.5808/GI.2017.15.4.170
HOTAIR Long Non-coding RNA: Characterizing the Locus Features by the In Silico Approaches
Abstract
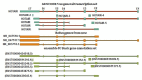
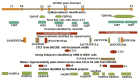
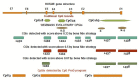
HOTAIR is an lncRNA that has been known to have an oncogenic role in different cancers. There is limited knowledge of genetic and epigenetic elements and their interactions for the gene encoding HOTAIR. Therefore, understanding the molecular mechanism and its regulation remains to be challenging. We used different in silico analyses to find genetic and epigenetic elements of HOTAIR gene to gain insight into its regulation. We reported different regulatory elements including canonical promoters, transcription start sites, CpGIs as well as epigenetic marks that are potentially involved in the regulation of HOTAIR gene expression. We identified repeat sequences and single nucleotide polymorphisms that are located within or next to the CpGIs of HOTAIR. Our analyses may help to find potential interactions between genetic and epigenetic elements of HOTAIR gene in the human tissues and show opportunities and limitations for researches on HOTAIR gene in future studies.
Keywords: CpG Islands; HOTAIR; bioinformatics; epigenetics; gene expression.
Figures
Similar articles
-
Characterizing the Retinoblastoma 1 locus: putative elements for Rb1 regulation by in silico analysis.Front Genet. 2014 Jan 28;5:2. doi: 10.3389/fgene.2014.00002. eCollection 2014. Front Genet. 2014. PMID: 24478791 Free PMC article.
-
HOTAIR: an oncogenic long non-coding RNA in different cancers.Cancer Biol Med. 2015 Mar;12(1):1-9. doi: 10.7497/j.issn.2095-3941.2015.0006. Cancer Biol Med. 2015. PMID: 25859406 Free PMC article. Review.
-
HOTAIR LncRNA: A novel oncogenic propellant in human cancer.Clin Chim Acta. 2020 Apr;503:1-18. doi: 10.1016/j.cca.2019.12.028. Epub 2019 Dec 31. Clin Chim Acta. 2020. PMID: 31901481 Review.
-
G-quadruplex is critical to epigenetic activation of the lncRNA HOTAIR in cancer cells.iScience. 2023 Nov 22;26(12):108559. doi: 10.1016/j.isci.2023.108559. eCollection 2023 Dec 15. iScience. 2023. PMID: 38144452 Free PMC article.
-
Quantitative Assessment of the Polymorphisms in the HOTAIR lncRNA and Cancer Risk: A Meta-Analysis of 8 Case-Control Studies.PLoS One. 2016 Mar 24;11(3):e0152296. doi: 10.1371/journal.pone.0152296. eCollection 2016. PLoS One. 2016. PMID: 27010768 Free PMC article.
Cited by
-
De novo annotation of lncRNA HOTAIR transcripts by long-read RNA capture-seq reveals a differentiation-driven isoform switch.BMC Genomics. 2022 Sep 17;23(1):658. doi: 10.1186/s12864-022-08887-w. BMC Genomics. 2022. PMID: 36115964 Free PMC article.
-
The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment.J Transl Med. 2020 Apr 3;18(1):152. doi: 10.1186/s12967-020-02320-0. J Transl Med. 2020. PMID: 32245498 Free PMC article. Review.
-
Deregulated Expression of Long Non-coding RNA HOX Transcript Antisense RNA (HOTAIR) in Egyptian Patients with Multiple Myeloma.Indian J Hematol Blood Transfus. 2020 Apr;36(2):271-276. doi: 10.1007/s12288-019-01211-9. Epub 2019 Oct 16. Indian J Hematol Blood Transfus. 2020. PMID: 32425377 Free PMC article.
-
Association Between SNPs of Long Non-coding RNA HOTAIR and Risk of Different Cancers.Front Genet. 2019 Feb 28;10:113. doi: 10.3389/fgene.2019.00113. eCollection 2019. Front Genet. 2019. PMID: 30873206 Free PMC article. No abstract available.
-
HOTAIR in colorectal cancer: structure, function, and therapeutic potential.Med Oncol. 2023 Aug 2;40(9):259. doi: 10.1007/s12032-023-02131-5. Med Oncol. 2023. PMID: 37530984 Review.
References
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. - PubMed
-
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. - PubMed
-
- Yu X, Li Z. Long non-coding RNA HOTAIR: a novel oncogene (review) Mol Med Rep. 2015;12:5611–5618. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
