Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing
- PMID: 29307488
- PMCID: PMC5766405
- DOI: 10.1016/j.cell.2017.12.007
Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing
Abstract
Ductal carcinoma in situ (DCIS) is an early-stage breast cancer that infrequently progresses to invasive ductal carcinoma (IDC). Genomic evolution has been difficult to delineate during invasion due to intratumor heterogeneity and the low number of tumor cells in the ducts. To overcome these challenges, we developed Topographic Single Cell Sequencing (TSCS) to measure genomic copy number profiles of single tumor cells while preserving their spatial context in tissue sections. We applied TSCS to 1,293 single cells from 10 synchronous patients with both DCIS and IDC regions in addition to exome sequencing. Our data reveal a direct genomic lineage between in situ and invasive tumor subpopulations and further show that most mutations and copy number aberrations evolved within the ducts prior to invasion. These results support a multiclonal invasion model, in which one or more clones escape the ducts and migrate into the adjacent tissues to establish the invasive carcinomas.
Keywords: breast cancer; breast cancer progression; cancer genomics; clonal evolution; copy number evolution; ductal carcinoma in situ; genome evolution; intratumor heterogeneity; invasion; single-cell sequencing.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



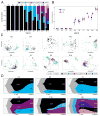

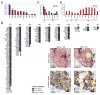

Comment in
-
On the trail of invasive cells in breast cancer.Nature. 2018 Feb 15;554(7692):308-309. doi: 10.1038/d41586-018-01634-w. Nature. 2018. PMID: 29446387 No abstract available.
References
-
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O’Connell P, Tsimelzon A, Medina D. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. - PubMed
-
- Andl CD, Mizushima T, Oyama K, Bowser M, Nakagawa H, Rustgi AK. EGFR-induced cell migration is mediated predominantly by the JAK-STAT pathway in primary esophageal keratinocytes. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1227–1237. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

