Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance
- PMID: 29307555
- PMCID: PMC5787036
- DOI: 10.1016/j.cub.2017.11.064
Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance
Abstract
The clearance of mitochondria by autophagy, mitophagy, is important for cell and organism health [1], and known to be regulated by ubiquitin. During Drosophila intestine development, cells undergo a dramatic reduction in cell size and clearance of mitochondria that depends on autophagy, the E1 ubiquitin-activating enzyme Uba1, and ubiquitin [2]. Here we screen a collection of putative ubiquitin-binding domain-encoding genes for cell size reduction and autophagy phenotypes. We identify the endosomal sorting complex required for transport (ESCRT) components TSG101 and Vps36, as well as the novel gene Vps13D. Vps13D is an essential gene that is necessary for autophagy, mitochondrial size, and mitochondrial clearance in Drosophila. Interestingly, a similar mitochondrial phenotype is observed in VPS13D mutant human cells. The ubiquitin-associated (UBA) domain of Vps13D binds K63 ubiquitin chains, and mutants lacking the UBA domain have defects in mitochondrial size and clearance and exhibit semi-lethality, highlighting the importance of Vps13D ubiquitin binding in both mitochondrial health and development. VPS13D mutant cells possess phosphorylated DRP1 and mitochondrial fission factor (MFF) as well as DRP1 association with mitochondria, suggesting that VPS13D functions downstream of these known regulators of mitochondrial fission. In addition, the large Vps13D mitochondrial and cell size phenotypes are suppressed by decreased mitochondrial fusion gene function. Thus, these results provide a previously unknown link between ubiquitin, mitochondrial size regulation, and autophagy.
Keywords: Drosophila; Vps13D; autophagy; fission; mitochondria; mitophagy; ubiquitin.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
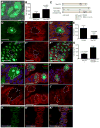
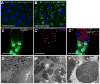
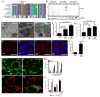

Comment in
-
Mitophagy: Vps13D Couples Mitochondrial Fission and Autophagic Clearance.Curr Biol. 2018 Jan 22;28(2):R66-R68. doi: 10.1016/j.cub.2017.12.017. Curr Biol. 2018. PMID: 29374445
References
-
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nature cell biology. 2014;16:495–501. - PubMed
-
- Rusten TE, Stenmark H. How do ESCRT proteins control autophagy? Journal of cell science. 2009;122:2179–2183. - PubMed
-
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Developmental cell. 2011;21:77–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

