A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle
- PMID: 29307657
- PMCID: PMC6172038
- DOI: 10.1016/j.jaci.2017.12.974
A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle
Abstract
Chromosome 17q12-21 remains the most highly replicated and significant asthma locus. Genotypes in the core region defined by the first genome-wide association study correlate with expression of 2 genes, ORM1-like 3 (ORMDL3) and gasdermin B (GSDMB), making these prime candidate asthma genes, although recent studies have implicated gasdermin A (GSDMA) distal to and post-GPI attachment to proteins 3 (PGAP3) proximal to the core region as independent loci. We review 10 years of studies on the 17q12-21 locus and suggest that genotype-specific risks for asthma at the proximal and distal loci are not specific to early-onset asthma and mediated by PGAP3, ORMDL3, and/or GSDMA expression. We propose that the weak and inconsistent associations of 17q single nucleotide polymorphisms with asthma in African Americans is due to the high frequency of some 17q alleles, the breakdown of linkage disequilibrium on African-derived chromosomes, and possibly different early-life asthma endotypes in these children. Finally, the inconsistent association between asthma and gene expression levels in blood or lung cells from older children and adults suggests that genotype effects may mediate asthma risk or protection during critical developmental windows and/or in response to relevant exposures in early life. Thus studies of young children and ethnically diverse populations are required to fully understand the relationship between genotype and asthma phenotype and the gene regulatory architecture at this locus.
Keywords: GSDMA; GSDMB; ORMDL3; PGAP3; Wheezing; gene expression; genome-wide association study; immune cells; lung cells.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
Figures


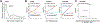
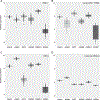
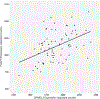

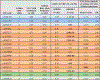


References
-
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

