Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties
- PMID: 29307839
- PMCID: PMC5856591
- DOI: 10.1016/j.chembiol.2017.12.007
Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties
Abstract
Invasive fungal infections are accompanied by high mortality rates that range up to 90%. At present, only three different compound classes are available for use in the clinic, and these often suffer from low bioavailability, toxicity, and drug resistance. These issues emphasize an urgent need for novel antifungal agents. Herein, we report the identification of chemically versatile benzamide and picolinamide scaffolds with antifungal properties. Chemogenomic profiling and biochemical assays with purified protein identified Sec14p, the major phosphatidylinositol/phosphatidylcholine transfer protein in Saccharomyces cerevisiae, as the sole essential target for these compounds. A functional variomics screen identified resistance-conferring residues that localized to the lipid-binding pocket of Sec14p. Determination of the X-ray co-crystal structure of a Sec14p-compound complex confirmed binding in this cavity and rationalized both the resistance-conferring residues and the observed structure-activity relationships. Taken together, these findings open new avenues for rational compound optimization and development of novel antifungal agents.
Keywords: Sec14p; antifungal; benzamide; chemogenomics; co-crystal; functional variomics; lipid-transfer protein; picolinamide; target identification.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors with the affiliation Novartis Institutes for BioMedical research are employees of Novartis Pharma AG and may own stock in the company. All other authors declare no competing financial interest.
Figures

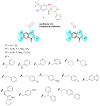
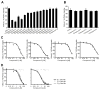

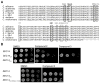
Comment in
-
Fungal Secretion: The Next-Gen Target of Antifungal Agents?Cell Chem Biol. 2018 Mar 15;25(3):233-235. doi: 10.1016/j.chembiol.2018.02.015. Cell Chem Biol. 2018. PMID: 29547713
References
-
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

