Combination of Ru(ii) complexes and light: new frontiers in cancer therapy
- PMID: 29308166
- PMCID: PMC5639435
- DOI: 10.1039/c4sc03759f
Combination of Ru(ii) complexes and light: new frontiers in cancer therapy
Abstract
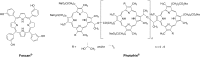
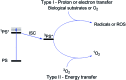
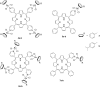
The synergistic action of light, oxygen and a photosensitizer (PS) has found applications for decades in medicine under the name of photodynamic therapy (PDT) for the treatment of skin diseases and, more recently, for the treatment of cancer. However, of the thirteen PSs currently approved for the treatment of cancer over more than 10 countries, only two contain a metal ion. This fact is rather surprising considering that nowadays around 50% of conventional chemotherapies involve the use of cisplatin and other platinum-containing drugs. In this perspective article, we review the opportunities brought by the use of Ru(ii) complexes as PSs in PDT. In addition, we also present the recent achievements in the application of Ru(ii) complexes in photoactivated chemotherapy (PACT). In this strategy, the presence of oxygen is not required to achieve cell toxicity. This is of significance since tumors are generally hypoxic. Importantly, this perspective article focuses particularly on the Ru(ii) complexes for which an in vitro biological evaluation has been performed and the mechanism of action (partially) unveiled.
Figures






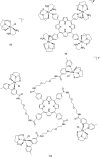
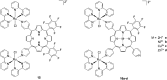

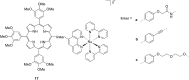
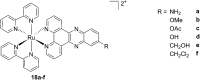
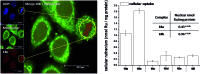
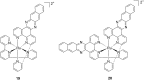
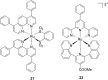
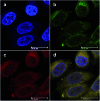

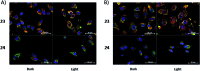

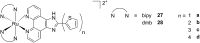

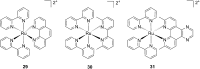
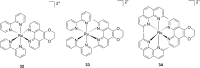
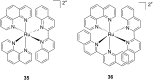



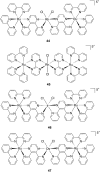

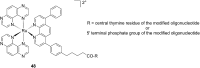
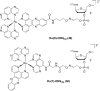

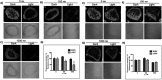
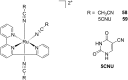
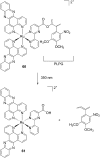

References
-
- Bergamo A., Gaiddon C., Schellens J. H. M., Beijnen J. H., Sava G. J. Inorg. Biochem. 2012;106:90–99. - PubMed
-
- Kilah N. L., Meggers E. Aust. J. Chem. 2012;65:1325–1332.
-
- Salassa L. Eur. J. Inorg. Chem. 2011:4931–4947.
-
- Sava G., Zorzet S., Turrin C., Vita F., Soranzo M., Zabucchi G., Cocchietto M., Bergamo A., DiGiovine S., Pezzoni G., Sartor L., Garbisa S. Clin. Cancer Res. 2003;9:1898–1905. - PubMed
-
- Rademaker-Lakhai J. M., van den Bongard D., Pluim D., Beijnen J. H., Schellens J. H. M. Clin. Cancer Res. 2004;10:3717–3727. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

