Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer
- PMID: 29308883
- PMCID: PMC5967631
- DOI: 10.1021/acs.biochem.7b01102
Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer
Abstract
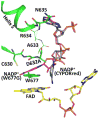
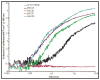
Conformational changes in NADPH-cytochrome P450 oxidoreductase (CYPOR) associated with electron transfer from NADPH to electron acceptors via FAD and FMN have been investigated via structural studies of the four-electron-reduced NADP+-bound enzyme and kinetic and structural studies of mutants that affect the conformation of the mobile Gly631-Asn635 loop (Asp632 loop). The structure of four-electron-reduced, NADP+-bound wild type CYPOR shows the plane of the nicotinamide ring positioned perpendicular to the FAD isoalloxazine with its carboxamide group forming H-bonds with N1 of the flavin ring and the Thr535 hydroxyl group. In the reduced enzyme, the C8-C8 atoms of the two flavin rings are ∼1 Å closer than in the fully oxidized and one-electron-reduced structures, which suggests that flavin reduction facilitates interflavin electron transfer. Structural and kinetic studies of mutants Asp632Ala, Asp632Phe, Asp632Asn, and Asp632Glu demonstrate that the carboxyl group of Asp632 is important for stabilizing the Asp632 loop in a retracted position that is required for the binding of the NADPH ribityl-nicotinamide in a hydride-transfer-competent conformation. Structures of the mutants and reduced wild type CYPOR permit us to identify a possible pathway for NADP(H) binding to and release from CYPOR. Asp632 mutants unable to form stable H-bonds with the backbone amides of Arg634, Asn635, and Met636 exhibit decreased catalytic activity and severely impaired hydride transfer from NADPH to FAD, but leave interflavin electron transfer intact. Intriguingly, the Arg634Ala mutation slightly increases the cytochrome P450 2B4 activity. We propose that Asp632 loop movement, in addition to facilitating NADP(H) binding and release, participates in domain movements modulating interflavin electron transfer.
Conflict of interest statement
Notes
The authors declare no competing financial interests.
Figures

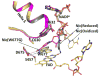









References
-
- Kim JJ, Waskell L. Electron Transfer Partners. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 4. Springer; Switzerland: 2015. pp. 33–68.
-
- Paine MJ, Scrutton NS, Munro AW, Gutierrez A, Roberts GCK, Wolf CR. Electron Transfer Partners of Cytochrome P450. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 3. Kluwer Academic/Plenum Publishers; New York, Boston, Dordrecht, London, Moscow: 2005. pp. 115–148.
-
- Guengerich FP. Human Cytochrome P450 Enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 3. Vol. 1. Kluwer Academic/Plenum Publishers; New York, NY: 2005. pp. 377–463.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

