CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation
- PMID: 29311295
- PMCID: PMC5789949
- DOI: 10.1073/pnas.1718059115
CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation
Abstract
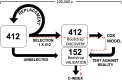
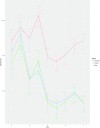
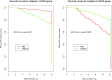
We evaluated the influence of an antioxidant and zinc nutritional supplement [the Age-Related Eye Disease Study (AREDS) formulation] on delaying or preventing progression to neovascular AMD (NV) in persons with age-related macular degeneration (AMD). AREDS subjects (n = 802) with category 3 or 4 AMD at baseline who had been treated with placebo or the AREDS formulation were evaluated for differences in the risk of progression to NV as a function of complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2) genotype groups. We used published genetic grouping: a two-SNP haplotype risk-calling algorithm to assess CFH, and either the single SNP rs10490924 or 372_815del443ins54 to mark ARMS2 risk. Progression risk was determined using the Cox proportional hazard model. Genetics-treatment interaction on NV risk was assessed using a multiiterative bootstrap validation analysis. We identified strong interaction of genetics with AREDS formulation treatment on the development of NV. Individuals with high CFH and no ARMS2 risk alleles and taking the AREDS formulation had increased progression to NV compared with placebo. Those with low CFH risk and high ARMS2 risk had decreased progression risk. Analysis of CFH and ARMS2 genotype groups from a validation dataset reinforces this conclusion. Bootstrapping analysis confirms the presence of a genetics-treatment interaction and suggests that individual treatment response to the AREDS formulation is largely determined by genetics. The AREDS formulation modifies the risk of progression to NV based on individual genetics. Its use should be based on patient-specific genotype.
Keywords: bootstrap validation; genetic effect modification; macular degeneration; ophthalmology; statistical interaction.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: B.W.Z. is the director of Arctic Medical Laboratories and founder and an equity holder of ArcticDx, Inc. (>5%), which owns patents relevant to the results; C.C.A. is a medical consultant and equity holder of ArcticAx Inc. (<1%); and R.K. is a technical consultant and equity holder of ArcticAx Inc. (<1%).
Figures


Comment in
-
Reply to Vickers: Pharmacogenetics and progression to neovascular age-related macular degeneration-Evidence supporting practice change.Proc Natl Acad Sci U S A. 2018 Jun 19;115(25):E5640-E5641. doi: 10.1073/pnas.1804781115. Epub 2018 Jun 7. Proc Natl Acad Sci U S A. 2018. PMID: 29880713 Free PMC article. No abstract available.
-
Pharmacogenomics of antioxidant supplementation to prevent age-related macular degeneration.Proc Natl Acad Sci U S A. 2018 Jun 19;115(25):E5639. doi: 10.1073/pnas.1803536115. Epub 2018 Jun 7. Proc Natl Acad Sci U S A. 2018. PMID: 29880714 Free PMC article. No abstract available.
References
-
- Schmier JK, Covert DW, Lau EC. Patterns and costs associated with progression of age-related macular degeneration. Am J Ophthalmol. 2012;154:675–681.e1. - PubMed
-
- Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. - PubMed
-
- Lechanteur YT, et al. Association of smoking and CFH and ARMS2 risk variants with younger age at onset of neovascular age-related macular degeneration. JAMA Ophthalmol. 2015;133:533–541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

