Physiological and Pathological Transcriptional Activation of Endogenous Retroelements Assessed by RNA-Sequencing of B Lymphocytes
- PMID: 29312197
- PMCID: PMC5733090
- DOI: 10.3389/fmicb.2017.02489
Physiological and Pathological Transcriptional Activation of Endogenous Retroelements Assessed by RNA-Sequencing of B Lymphocytes
Abstract
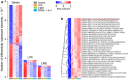
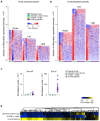
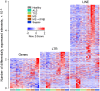
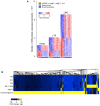
In addition to evolutionarily-accrued sequence mutation or deletion, endogenous retroelements (EREs) in eukaryotic genomes are subject to epigenetic silencing, preventing or reducing their transcription, particularly in the germplasm. Nevertheless, transcriptional activation of EREs, including endogenous retroviruses (ERVs) and long interspersed nuclear elements (LINEs), is observed in somatic cells, variably upon cellular differentiation and frequently upon cellular transformation. ERE transcription is modulated during physiological and pathological immune cell activation, as well as in immune cell cancers. However, our understanding of the potential consequences of such modulation remains incomplete, partly due to the relative scarcity of information regarding genome-wide ERE transcriptional patterns in immune cells. Here, we describe a methodology that allows probing RNA-sequencing (RNA-seq) data for genome-wide expression of EREs in murine and human cells. Our analysis of B cells reveals that their transcriptional response during immune activation is dominated by induction of gene transcription, and that EREs respond to a much lesser extent. The transcriptional activity of the majority of EREs is either unaffected or reduced by B cell activation both in mice and humans, albeit LINEs appear considerably more responsive in the latter host. Nevertheless, a small number of highly distinct ERVs are strongly and consistently induced during B cell activation. Importantly, this pattern contrasts starkly with B cell transformation, which exhibits widespread induction of EREs, including ERVs that minimally overlap with those responsive to immune stimulation. The distinctive patterns of ERE induction suggest different underlying mechanisms and will help separate physiological from pathological expression.
Keywords: B cell lymphoma; B lymphocyte activation; autoimmunity; cancer; endogenous retroelements; endogenous retroviruses; genetic; transcription.
Figures


References
-
- Brudek T., Christensen T., Aagaard L., Petersen T., Hansen H. J., Moller-Larsen A. (2009). B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 6:104. 10.1186/1742-4690-6-104 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

