Arginine methylation of EGFR: a new biomarker for predicting resistance to anti-EGFR treatment
- PMID: 29312811
- PMCID: PMC5752698
Arginine methylation of EGFR: a new biomarker for predicting resistance to anti-EGFR treatment
Abstract
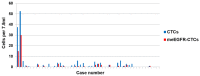
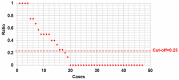
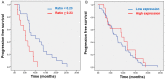
Arginine methylation of the epidermal growth factor receptor (meEGFR) increases the binding affinity of EGFR ligands and is reported to have a role in predicting response to anti-EGFR agents. This study investigated the predictive impact of meEGFR in metastatic colorectal cancer (mCRC) patients treated with anti-EGFR agents. Two patient cohorts were evaluated. Cohort 1 consisted of mCRC patients with documented disease progression following anti-EGFR treatment. Circulating tumor cells (CTCs) were isolated and distinguished based on CD45- and Epcam+. Cohort 2 consisted of formalin fixed paraffin-embedded (FFPE) blocks from a prospective cohort. meEGFR in both cohorts was identified by positive staining for me-R198/200 EGFR signal. CTCs were identified in 30 out of 47 cases in cohort 1. Of those 30, meEGFR-CTCs were identified in 19 cases. Mean total meEGFR-CTCs counts was 2.3 (range 0-30) cells per 7.5 ml. There was no association between meEGFR-CTCs and clinic-pathological-molecular features. In RASwt/BRAFwt patients with high levels of meEGFR-CTCs ratio (≥ 0.23) had significantly inferior PFS with anti-EGFR treatment (HR = 3.4, 95% CI 1.5-7.9, P = 0.004). By contrast, high levels of meEGFR in the untreated tumor tissues had no correlation with anti-EGFR treatment duration in cohort 2. Therefore, meEGFR-CTCs may have the potential to serve as a "liquid biopsy" biomarker to predict anti-EGFR treatment efficacy.
Keywords: EGFR; Liquid biopsy; arginine methylation; circulating tumor cells; colorectal cancer; predictive marker.
Conflict of interest statement
None.
Figures
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. - PubMed
-
- Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. - PubMed
-
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. - PubMed
-
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. - PubMed
-
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–72. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
