Human dendritic cell subsets: an update
- PMID: 29313948
- PMCID: PMC5904714
- DOI: 10.1111/imm.12888
Human dendritic cell subsets: an update
Abstract
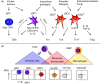
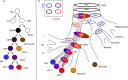
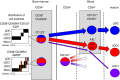
Dendritic cells (DC) are a class of bone-marrow-derived cells arising from lympho-myeloid haematopoiesis that form an essential interface between the innate sensing of pathogens and the activation of adaptive immunity. This task requires a wide range of mechanisms and responses, which are divided between three major DC subsets: plasmacytoid DC (pDC), myeloid/conventional DC1 (cDC1) and myeloid/conventional DC2 (cDC2). Each DC subset develops under the control of a specific repertoire of transcription factors involving differential levels of IRF8 and IRF4 in collaboration with PU.1, ID2, E2-2, ZEB2, KLF4, IKZF1 and BATF3. DC haematopoiesis is conserved between mammalian species and is distinct from monocyte development. Although monocytes can differentiate into DC, especially during inflammation, most quiescent tissues contain significant resident populations of DC lineage cells. An extended range of surface markers facilitates the identification of specific DC subsets although it remains difficult to dissociate cDC2 from monocyte-derived DC in some settings. Recent studies based on an increasing level of resolution of phenotype and gene expression have identified pre-DC in human blood and heterogeneity among cDC2. These advances facilitate the integration of mouse and human immunology, support efforts to unravel human DC function in vivo and continue to present new translational opportunities to medicine.
Keywords: antigen presentation/processing; dendritic cell; transcriptomics.
© 2018 The Authors. Immunology Published by John Wiley & Sons Ltd.
Figures

References
-
- Heidkamp GF, Sander J, Lehmann CHK, Heger L, Eissing N, Baranska A et al Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci Immunol 2016; 1:eaai7677. - PubMed
-
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S et al BDCA‐2, BDCA‐3, and BDCA‐4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol 2000; 165:6037–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

