HTT-DB: new features and updates
- PMID: 29315358
- PMCID: PMC7206651
- DOI: 10.1093/database/bax102
HTT-DB: new features and updates
Abstract
Horizontal Transfer (HT) of genetic material between species is a common phenomenon among Bacteria and Archaea species and several databases are available for information retrieval and data mining. However, little attention has been given to this phenomenon among eukaryotic species mainly due to the lower proportion of these events. In the last years, a vertiginous amount of new HT events involving eukaryotic species was reported in the literature, highlighting the need of a common repository to keep the scientific community up to date and describe overall trends. Recently, we published the first HT database focused on HT of transposable elements among eukaryotes: the Horizontal Transposon Transfer DataBase (http://lpa.saogabriel.unipampa.edu.br: 8080/httdatabase/). Here, we present new features and updates of this unique database: (i) its expansion to include virus-host exchange of genetic material, which we called Horizontal Virus Transfer (HVT) and (ii) the availability of a web server for HT detection, where we implemented the online version of vertical and horizontal inheritance consistence analysis (VHICA), an R package developed for HT detection. These improvements will help researchers to navigate through known HVT cases, take data-informed decision and export figures based on keywords searches. Moreover, the availability of the VHICA as an online tool will make this software easily reachable even for researchers with no or little computation knowledge as well as foster our capability to detect new HT events in a wide variety of taxa. (Database URL: http://lpa.saogabriel.unipampa.edu.br:8080/httdatabase/).
© The Author(s) 2018. Published by Oxford University Press.
Figures
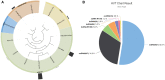

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

