Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility
- PMID: 29315591
- PMCID: PMC5873385
- DOI: 10.1111/nph.14970
Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility
Abstract
Feruloylation of arabinoxylan (AX) in grass cell walls is a key determinant of recalcitrance to enzyme attack, making it a target for improvement of grass crops, and of interest in grass evolution. Definitive evidence on the genes responsible is lacking so we studied a candidate gene that we identified within the BAHD acyl-CoA transferase family. We used RNA interference (RNAi) silencing of orthologs in the model grasses Setaria viridis (SvBAHD01) and Brachypodium distachyon (BdBAHD01) and determined effects on AX feruloylation. Silencing of SvBAHD01 in Setaria resulted in a c. 60% decrease in AX feruloylation in stems consistently across four generations. Silencing of BdBAHD01 in Brachypodium stems decreased feruloylation much less, possibly due to higher expression of functionally redundant genes. Setaria SvBAHD01 RNAi plants showed: no decrease in total lignin, approximately doubled arabinose acylated by p-coumarate, changes in two-dimensional NMR spectra of unfractionated cell walls consistent with biochemical estimates, no effect on total biomass production and an increase in biomass saccharification efficiency of 40-60%. We provide the first strong evidence for a key role of the BAHD01 gene in AX feruloylation and demonstrate that it is a promising target for improvement of grass crops for biofuel, biorefining and animal nutrition applications.
Keywords: cell wall acylation; ferulic acid; grass evolution; hydroxycinnamates; lignocellulosic feedstock.
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures

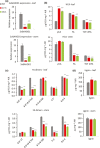
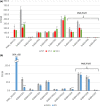
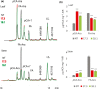

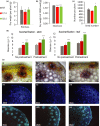
Comment in
-
Bringing down the wall one brick at a time.New Phytol. 2018 Apr;218(1):5-7. doi: 10.1111/nph.15052. New Phytol. 2018. PMID: 29488282 No abstract available.
References
-
- Akin DE. 1990. Diazonium compounds localize grass cell wall phenolics: relation to wall digestibility. Crop Science 30: 985–989.
-
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J et al 2012. Reference genome sequence of the model plant Setaria . Nature Biotechnology 30: 555–561. - PubMed
-
- Bouvier d'Yvoire M, Bouchabke‐Coussa O, Voorend W, Antelme S, Cézard L, Legée F, Lebris P, Legay S, Whitehead C, McQueen‐Mason SJ et al 2013. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon . Plant Journal 73: 496–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

