Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets
- PMID: 29316443
- PMCID: PMC5764114
- DOI: 10.1016/j.devcel.2017.12.011
Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets
Abstract
Cytosolic lipid droplets (LDs) are the main storage organelles for metabolic energy in most cells. They are unusual organelles that are bounded by a phospholipid monolayer and specific surface proteins, including key enzymes of lipid and energy metabolism. Proteins targeting LDs from the cytoplasm often contain amphipathic helices, but how they bind to LDs is not well understood. Combining computer simulations with experimental studies in vitro and in cells, we uncover a general mechanism for targeting of cytosolic proteins to LDs: large hydrophobic residues of amphipathic helices detect and bind to large, persistent membrane packing defects that are unique to the LD surface. Surprisingly, amphipathic helices with large hydrophobic residues from many different proteins are capable of binding to LDs. This suggests that LD protein composition is additionally determined by mechanisms that selectively prevent proteins from binding LDs, such as macromolecular crowding at the LD surface.
Keywords: all-atom molecular dynamics simulations; amphipathic helices; cell biology; lipid droplets; phospholipid bilayers; phospholipid monolayers; phospholipid packing defects; protein targeting; reconstitution assay.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
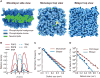
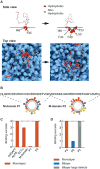
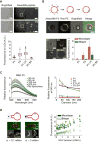
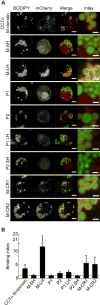

Comment in
-
Understanding the Lipid Droplet Proteome and Protein Targeting.Dev Cell. 2018 Jan 8;44(1):1-2. doi: 10.1016/j.devcel.2017.12.017. Epub 2018 Jan 8. Dev Cell. 2018. PMID: 29316437 Free PMC article.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;(1–2):19–25.
-
- Adrover M, Martorell G, Martin SR, Urosev D, Konarev PV, Svergun DI, Daura X, Temussi P, Pastore A. The role of hydration in protein stability: comparison of the cold and heat unfolded states of Yfh1. J Mol Biol. 2012;417:413–424. - PubMed
-
- Angelova MI, Soléau S, Méléard P, Faucon JF, Bothorel P. Preparation of giant vesicles by external AC electric fields . Kinetics and applications. Progress in Colloid & Polymer Science. 1992;89:127–131.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

